Magnesium in PDB 7mqu: The Haddock Model of Gdp Kras in Complex with Promethazine Using uc(Nmr) Chemical Shift Perturbations
Enzymatic activity of The Haddock Model of Gdp Kras in Complex with Promethazine Using uc(Nmr) Chemical Shift Perturbations
All present enzymatic activity of The Haddock Model of Gdp Kras in Complex with Promethazine Using uc(Nmr) Chemical Shift Perturbations:
3.6.5.2;
3.6.5.2;
Magnesium Binding Sites:
The binding sites of Magnesium atom in the The Haddock Model of Gdp Kras in Complex with Promethazine Using uc(Nmr) Chemical Shift Perturbations
(pdb code 7mqu). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total only one binding site of Magnesium was determined in the The Haddock Model of Gdp Kras in Complex with Promethazine Using uc(Nmr) Chemical Shift Perturbations, PDB code: 7mqu:
In total only one binding site of Magnesium was determined in the The Haddock Model of Gdp Kras in Complex with Promethazine Using uc(Nmr) Chemical Shift Perturbations, PDB code: 7mqu:
Magnesium binding site 1 out of 1 in 7mqu
Go back to
Magnesium binding site 1 out
of 1 in the The Haddock Model of Gdp Kras in Complex with Promethazine Using uc(Nmr) Chemical Shift Perturbations
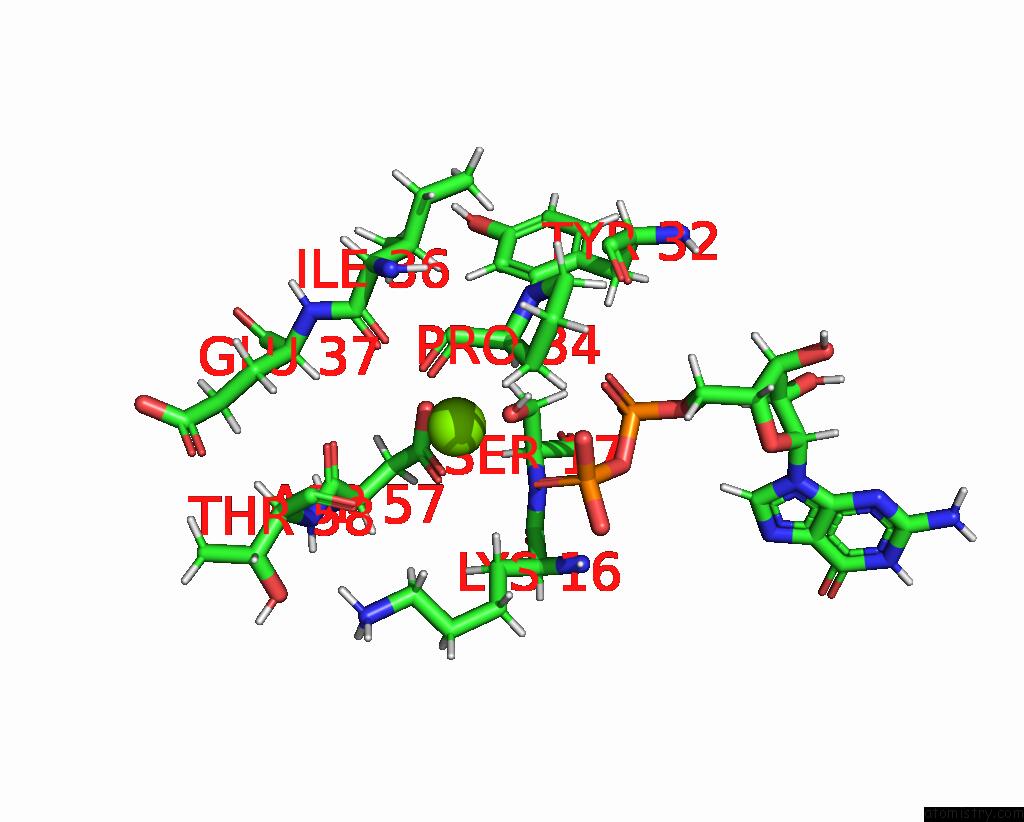
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of The Haddock Model of Gdp Kras in Complex with Promethazine Using uc(Nmr) Chemical Shift Perturbations within 5.0Å range:
|
Reference:
X.Wang,
A.A.Gorfe,
J.A.Putkey.
Antipsychotic Phenothiazine Drugs Bind to Kras in Vitro. J.Biomol.uc(Nmr) V. 75 233 2021.
ISSN: ISSN 0925-2738
PubMed: 34176062
DOI: 10.1007/S10858-021-00371-Z
Page generated: Thu Oct 3 01:05:46 2024
ISSN: ISSN 0925-2738
PubMed: 34176062
DOI: 10.1007/S10858-021-00371-Z
Last articles
K in 9G9VK in 9DTR
K in 9C46
K in 9G9W
K in 9G9X
K in 9ESI
K in 9ESH
K in 8ZEX
K in 8VAV
K in 8VAZ