Magnesium in PDB 8te0: 16MER Self-Complementary Duplex Rna with D:C Pair Sequence 1
Protein crystallography data
The structure of 16MER Self-Complementary Duplex Rna with D:C Pair Sequence 1, PDB code: 8te0
was solved by
Z.Fang,
X.Jia,
J.W.Szostak,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 35.41 / 1.42 |
| Space group | H 3 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 42.501, 42.501, 129.967, 90, 90, 120 |
| R / Rfree (%) | 21.5 / 23.8 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the 16MER Self-Complementary Duplex Rna with D:C Pair Sequence 1
(pdb code 8te0). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total only one binding site of Magnesium was determined in the 16MER Self-Complementary Duplex Rna with D:C Pair Sequence 1, PDB code: 8te0:
In total only one binding site of Magnesium was determined in the 16MER Self-Complementary Duplex Rna with D:C Pair Sequence 1, PDB code: 8te0:
Magnesium binding site 1 out of 1 in 8te0
Go back to
Magnesium binding site 1 out
of 1 in the 16MER Self-Complementary Duplex Rna with D:C Pair Sequence 1
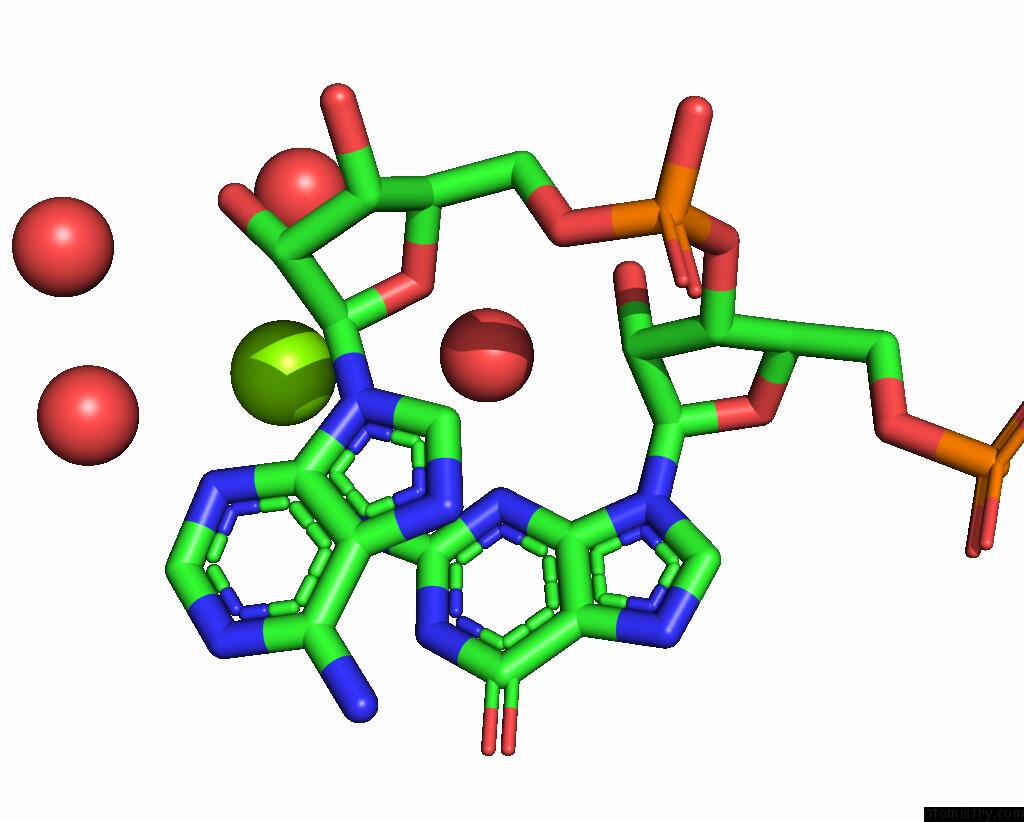
Mono view
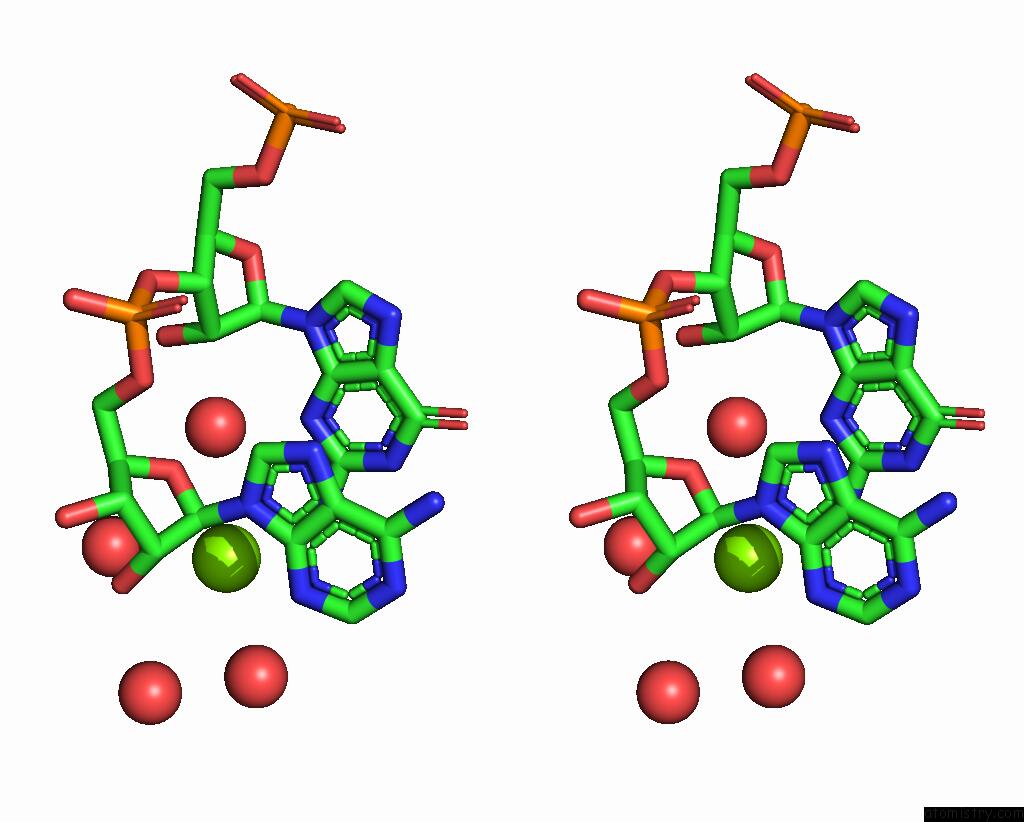
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of 16MER Self-Complementary Duplex Rna with D:C Pair Sequence 1 within 5.0Å range:
|
Reference:
X.Jia,
Z.Fang,
S.C.Kim,
D.Ding,
L.Zhou,
J.W.Szostak.
Diaminopurine in Nonenzymatic Rna Template Copying. J.Am.Chem.Soc. 2024.
ISSN: ESSN 1520-5126
PubMed: 38818863
DOI: 10.1021/JACS.4C02560
Page generated: Fri Oct 4 20:36:59 2024
ISSN: ESSN 1520-5126
PubMed: 38818863
DOI: 10.1021/JACS.4C02560
Last articles
Fe in 9DEUFe in 9CCB
Fe in 9D86
Fe in 9DCO
Fe in 9EBM
Fe in 9EBK
Fe in 8ZQD
Fe in 8ZEH
Fe in 8ZET
Fe in 8Z11