Magnesium in PDB 8tzj: Cryo-Em Structure of Vibrio Cholerae Ftse/Ftsx Complex
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Cryo-Em Structure of Vibrio Cholerae Ftse/Ftsx Complex
(pdb code 8tzj). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 2 binding sites of Magnesium where determined in the Cryo-Em Structure of Vibrio Cholerae Ftse/Ftsx Complex, PDB code: 8tzj:
Jump to Magnesium binding site number: 1; 2;
In total 2 binding sites of Magnesium where determined in the Cryo-Em Structure of Vibrio Cholerae Ftse/Ftsx Complex, PDB code: 8tzj:
Jump to Magnesium binding site number: 1; 2;
Magnesium binding site 1 out of 2 in 8tzj
Go back to
Magnesium binding site 1 out
of 2 in the Cryo-Em Structure of Vibrio Cholerae Ftse/Ftsx Complex
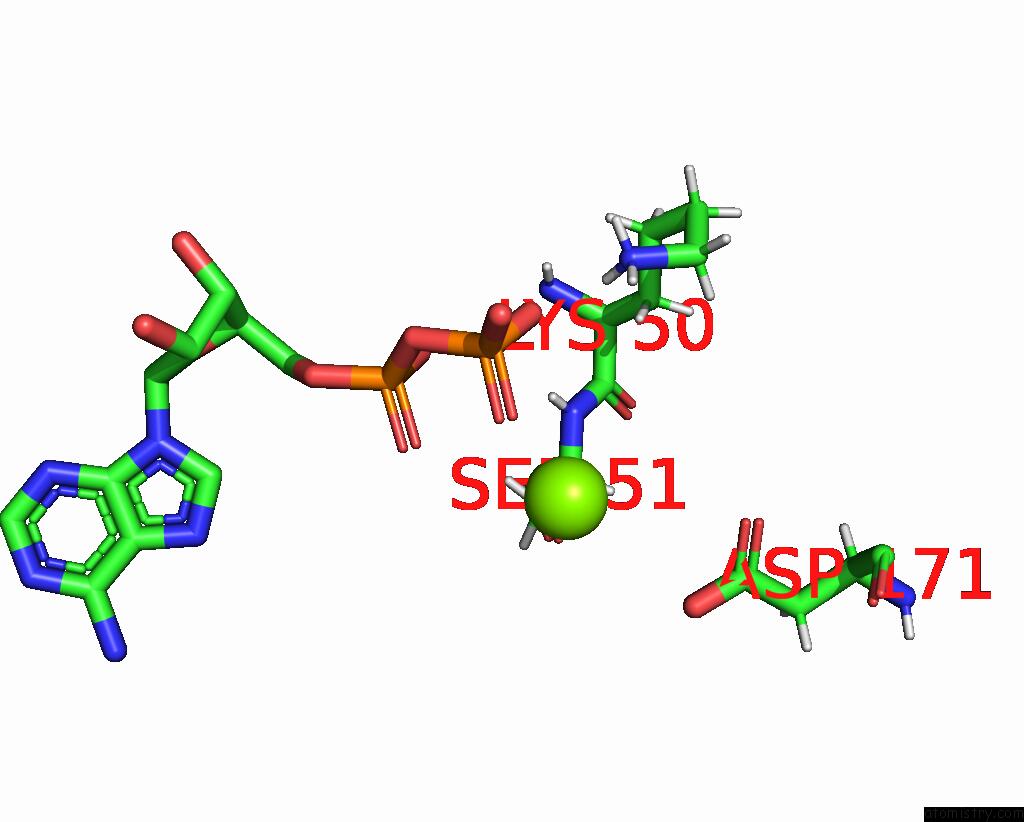
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Cryo-Em Structure of Vibrio Cholerae Ftse/Ftsx Complex within 5.0Å range:
|
Magnesium binding site 2 out of 2 in 8tzj
Go back to
Magnesium binding site 2 out
of 2 in the Cryo-Em Structure of Vibrio Cholerae Ftse/Ftsx Complex
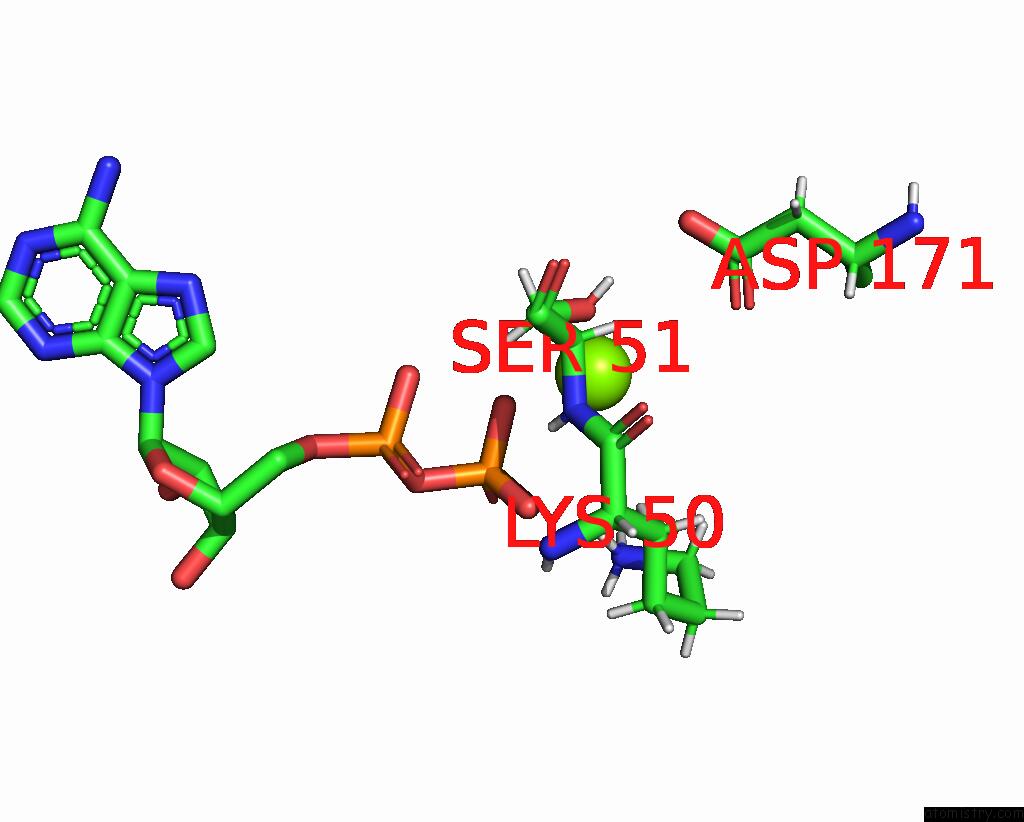
Mono view
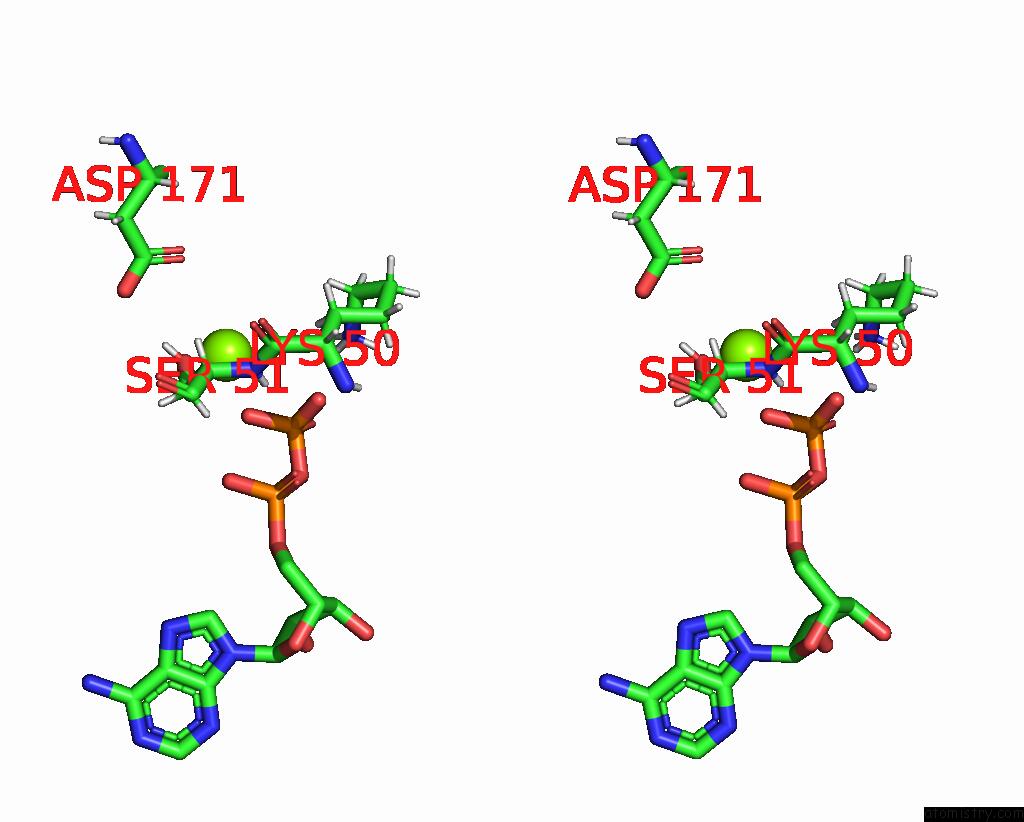
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Cryo-Em Structure of Vibrio Cholerae Ftse/Ftsx Complex within 5.0Å range:
|
Reference:
A.Hao,
Y.Suo,
S.Y.Lee.
Structural Insights Into the Ftsex-Envc Complex Regulation on Septal Peptidoglycan Hydrolysis in Vibrio Cholerae. Structure 2023.
ISSN: ISSN 0969-2126
PubMed: 38070498
DOI: 10.1016/J.STR.2023.11.007
Page generated: Fri Oct 4 21:07:48 2024
ISSN: ISSN 0969-2126
PubMed: 38070498
DOI: 10.1016/J.STR.2023.11.007
Last articles
I in 9AXJHg in 9D67
Hg in 9D66
Hg in 9D69
Hg in 9D6A
Fe in 9J2F
Fe in 9JDC
Fe in 9JDB
Fe in 9KU6
Fe in 9F47