Magnesium »
PDB 1ao0-1bh2 »
1bh2 »
Magnesium in PDB 1bh2: A326S Mutant of An Inhibitory Alpha Subunit
Protein crystallography data
The structure of A326S Mutant of An Inhibitory Alpha Subunit, PDB code: 1bh2
was solved by
M.B.Mixon,
B.A.Posner,
M.A.Wall,
A.G.Gilman,
S.R.Sprang,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 18.00 / 2.10 |
| Space group | P 32 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 79.584, 79.584, 105.556, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 19 / 25 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the A326S Mutant of An Inhibitory Alpha Subunit
(pdb code 1bh2). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total only one binding site of Magnesium was determined in the A326S Mutant of An Inhibitory Alpha Subunit, PDB code: 1bh2:
In total only one binding site of Magnesium was determined in the A326S Mutant of An Inhibitory Alpha Subunit, PDB code: 1bh2:
Magnesium binding site 1 out of 1 in 1bh2
Go back to
Magnesium binding site 1 out
of 1 in the A326S Mutant of An Inhibitory Alpha Subunit
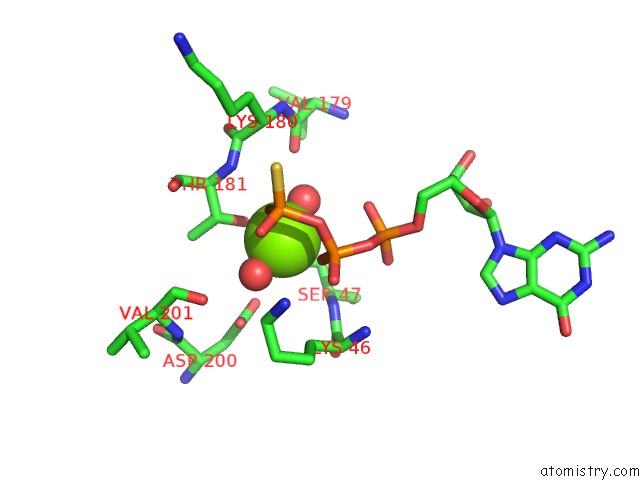
Mono view
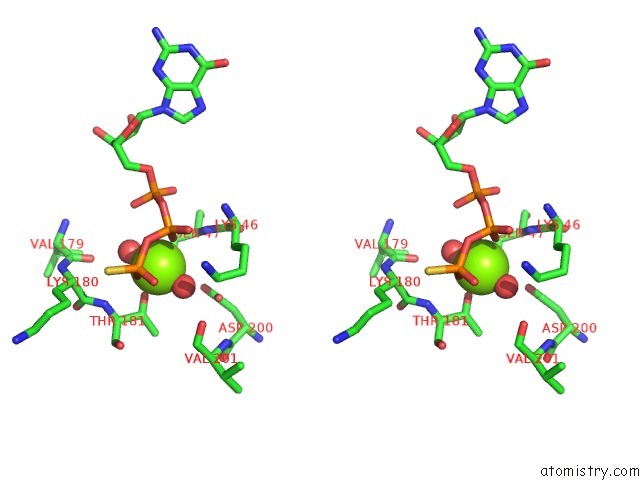
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of A326S Mutant of An Inhibitory Alpha Subunit within 5.0Å range:
|
Reference:
B.A.Posner,
M.B.Mixon,
M.A.Wall,
S.R.Sprang,
A.G.Gilman.
The A326S Mutant of GIALPHA1 As An Approximation of the Receptor-Bound State. J.Biol.Chem. V. 273 21752 1998.
ISSN: ISSN 0021-9258
PubMed: 9705312
DOI: 10.1074/JBC.273.34.21752
Page generated: Sat Aug 9 20:11:59 2025
ISSN: ISSN 0021-9258
PubMed: 9705312
DOI: 10.1074/JBC.273.34.21752
Last articles
Mg in 3CF5Mg in 3CCV
Mg in 3CCU
Mg in 3CCS
Mg in 3CCR
Mg in 3CCQ
Mg in 3CCM
Mg in 3CCL
Mg in 3CCJ
Mg in 3CC2