Magnesium »
PDB 1it7-1jbz »
1jah »
Magnesium in PDB 1jah: H-Ras P21 Protein Mutant G12P, Complexed with Guanosine-5'- [Beta,Gamma-Methylene] Triphosphate and Magnesium
Protein crystallography data
The structure of H-Ras P21 Protein Mutant G12P, Complexed with Guanosine-5'- [Beta,Gamma-Methylene] Triphosphate and Magnesium, PDB code: 1jah
was solved by
T.Schweins,
K.Scheffzek,
R.Assheuer,
A.Wittinghofer,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 8.00 / 1.80 |
| Space group | P 32 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 40.100, 40.100, 160.640, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 22 / 29 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the H-Ras P21 Protein Mutant G12P, Complexed with Guanosine-5'- [Beta,Gamma-Methylene] Triphosphate and Magnesium
(pdb code 1jah). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total only one binding site of Magnesium was determined in the H-Ras P21 Protein Mutant G12P, Complexed with Guanosine-5'- [Beta,Gamma-Methylene] Triphosphate and Magnesium, PDB code: 1jah:
In total only one binding site of Magnesium was determined in the H-Ras P21 Protein Mutant G12P, Complexed with Guanosine-5'- [Beta,Gamma-Methylene] Triphosphate and Magnesium, PDB code: 1jah:
Magnesium binding site 1 out of 1 in 1jah
Go back to
Magnesium binding site 1 out
of 1 in the H-Ras P21 Protein Mutant G12P, Complexed with Guanosine-5'- [Beta,Gamma-Methylene] Triphosphate and Magnesium
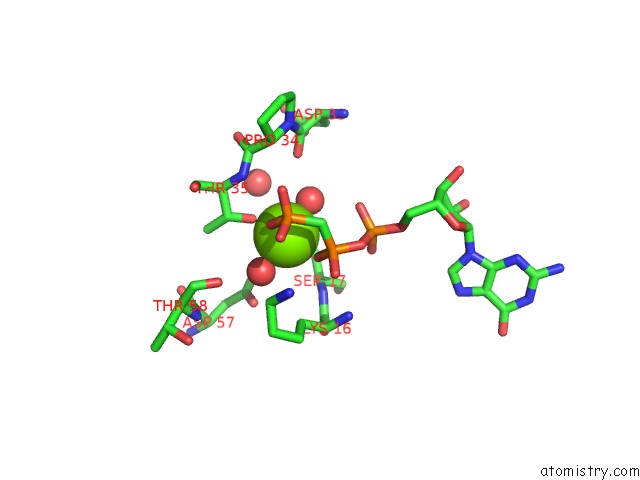
Mono view
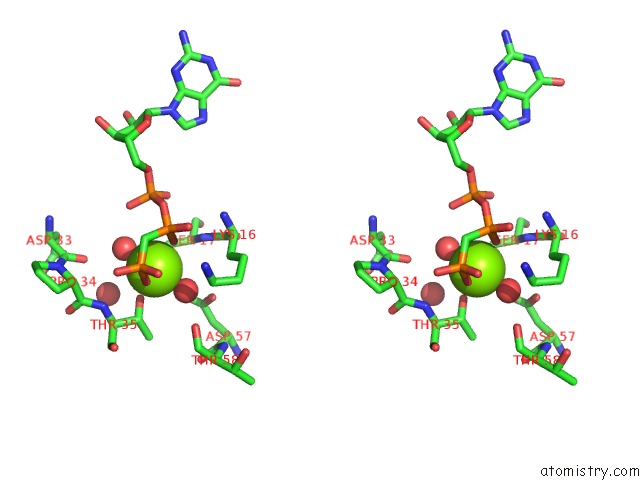
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of H-Ras P21 Protein Mutant G12P, Complexed with Guanosine-5'- [Beta,Gamma-Methylene] Triphosphate and Magnesium within 5.0Å range:
|
Reference:
T.Schweins,
K.Scheffzek,
R.Assheuer,
A.Wittinghofer.
The Role of the Metal Ion in the P21RAS Catalysed Gtp-Hydrolysis: MN2+ Versus MG2+. J.Mol.Biol. V. 266 847 1997.
ISSN: ISSN 0022-2836
PubMed: 9102473
DOI: 10.1006/JMBI.1996.0814
Page generated: Tue Aug 13 05:47:31 2024
ISSN: ISSN 0022-2836
PubMed: 9102473
DOI: 10.1006/JMBI.1996.0814
Last articles
F in 4LOQF in 4LQG
F in 4LPB
F in 4LOP
F in 4LP0
F in 4LOY
F in 4LMN
F in 4LOO
F in 4LN7
F in 4LE0