Magnesium »
PDB 1l3r-1lny »
1l5u »
Magnesium in PDB 1l5u: Crystal Structure of Bacillus Dna Polymerase I Fragment Product Complex with 12 Base Pairs of Duplex Dna Following Addition of A Dttp, A Datp, and A Dctp Residue.
Enzymatic activity of Crystal Structure of Bacillus Dna Polymerase I Fragment Product Complex with 12 Base Pairs of Duplex Dna Following Addition of A Dttp, A Datp, and A Dctp Residue.
All present enzymatic activity of Crystal Structure of Bacillus Dna Polymerase I Fragment Product Complex with 12 Base Pairs of Duplex Dna Following Addition of A Dttp, A Datp, and A Dctp Residue.:
2.7.7.7;
2.7.7.7;
Protein crystallography data
The structure of Crystal Structure of Bacillus Dna Polymerase I Fragment Product Complex with 12 Base Pairs of Duplex Dna Following Addition of A Dttp, A Datp, and A Dctp Residue., PDB code: 1l5u
was solved by
S.J.Johnson,
J.S.Taylor,
L.S.Beese,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 33.69 / 1.95 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 87.469, 93.418, 105.670, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 19.5 / 22.9 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Crystal Structure of Bacillus Dna Polymerase I Fragment Product Complex with 12 Base Pairs of Duplex Dna Following Addition of A Dttp, A Datp, and A Dctp Residue.
(pdb code 1l5u). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total only one binding site of Magnesium was determined in the Crystal Structure of Bacillus Dna Polymerase I Fragment Product Complex with 12 Base Pairs of Duplex Dna Following Addition of A Dttp, A Datp, and A Dctp Residue., PDB code: 1l5u:
In total only one binding site of Magnesium was determined in the Crystal Structure of Bacillus Dna Polymerase I Fragment Product Complex with 12 Base Pairs of Duplex Dna Following Addition of A Dttp, A Datp, and A Dctp Residue., PDB code: 1l5u:
Magnesium binding site 1 out of 1 in 1l5u
Go back to
Magnesium binding site 1 out
of 1 in the Crystal Structure of Bacillus Dna Polymerase I Fragment Product Complex with 12 Base Pairs of Duplex Dna Following Addition of A Dttp, A Datp, and A Dctp Residue.
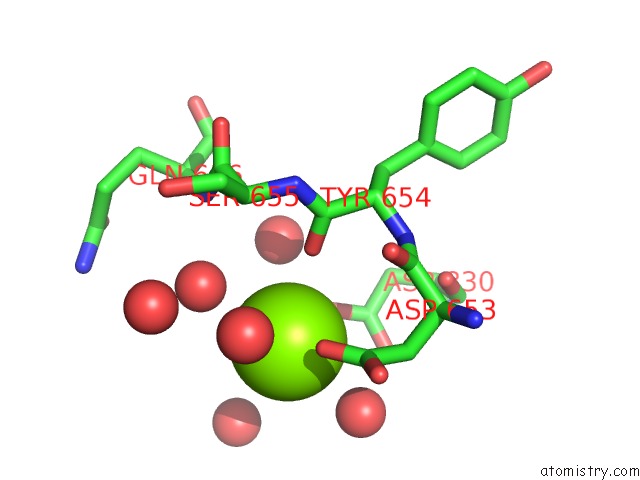
Mono view
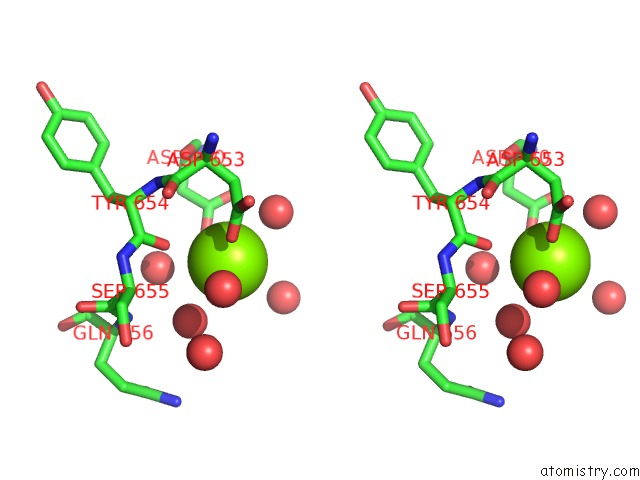
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Crystal Structure of Bacillus Dna Polymerase I Fragment Product Complex with 12 Base Pairs of Duplex Dna Following Addition of A Dttp, A Datp, and A Dctp Residue. within 5.0Å range:
|
Reference:
S.J.Johnson,
J.S.Taylor,
L.S.Beese.
Processive Dna Synthesis Observed in A Polymerase Crystal Suggests A Mechanism For the Prevention of Frameshift Mutations Proc.Natl.Acad.Sci.Usa V. 100 3895 2003.
ISSN: ISSN 0027-8424
PubMed: 12649320
DOI: 10.1073/PNAS.0630532100
Page generated: Sun Aug 10 00:38:00 2025
ISSN: ISSN 0027-8424
PubMed: 12649320
DOI: 10.1073/PNAS.0630532100
Last articles
Mg in 4DUZMg in 4DUY
Mg in 4DR7
Mg in 4DR6
Mg in 4DR5
Mg in 4DUX
Mg in 4DUW
Mg in 4DUV
Mg in 4DUO
Mg in 4DUG