Magnesium »
PDB 1ozf-1php »
1p18 »
Magnesium in PDB 1p18: Hypoxanthine Phosphoribosyltransferase From Trypanosoma Cruzi, K68R Mutant, Ternary Substrates Complex
Enzymatic activity of Hypoxanthine Phosphoribosyltransferase From Trypanosoma Cruzi, K68R Mutant, Ternary Substrates Complex
All present enzymatic activity of Hypoxanthine Phosphoribosyltransferase From Trypanosoma Cruzi, K68R Mutant, Ternary Substrates Complex:
2.4.2.8;
2.4.2.8;
Protein crystallography data
The structure of Hypoxanthine Phosphoribosyltransferase From Trypanosoma Cruzi, K68R Mutant, Ternary Substrates Complex, PDB code: 1p18
was solved by
B.Canyuk,
A.E.Eakin,
S.P.Craig Iii,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 20.00 / 2.00 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 39.500, 102.100, 51.800, 90.00, 94.30, 90.00 |
| R / Rfree (%) | 21 / 29.3 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Hypoxanthine Phosphoribosyltransferase From Trypanosoma Cruzi, K68R Mutant, Ternary Substrates Complex
(pdb code 1p18). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 4 binding sites of Magnesium where determined in the Hypoxanthine Phosphoribosyltransferase From Trypanosoma Cruzi, K68R Mutant, Ternary Substrates Complex, PDB code: 1p18:
Jump to Magnesium binding site number: 1; 2; 3; 4;
In total 4 binding sites of Magnesium where determined in the Hypoxanthine Phosphoribosyltransferase From Trypanosoma Cruzi, K68R Mutant, Ternary Substrates Complex, PDB code: 1p18:
Jump to Magnesium binding site number: 1; 2; 3; 4;
Magnesium binding site 1 out of 4 in 1p18
Go back to
Magnesium binding site 1 out
of 4 in the Hypoxanthine Phosphoribosyltransferase From Trypanosoma Cruzi, K68R Mutant, Ternary Substrates Complex
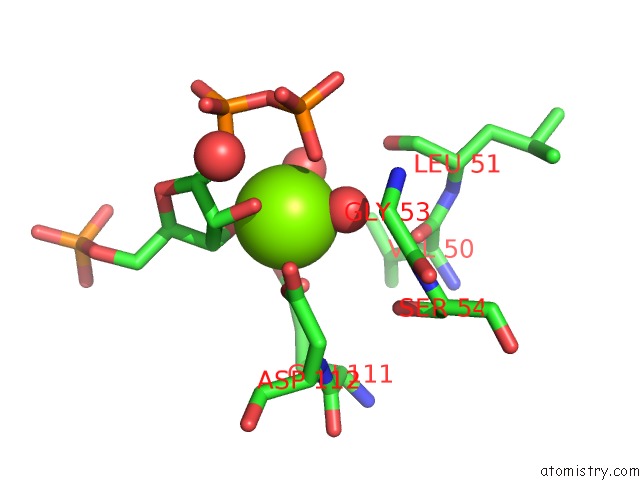
Mono view
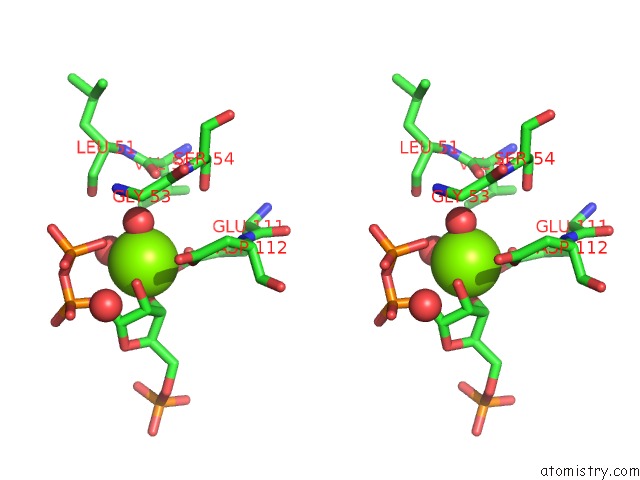
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Hypoxanthine Phosphoribosyltransferase From Trypanosoma Cruzi, K68R Mutant, Ternary Substrates Complex within 5.0Å range:
|
Magnesium binding site 2 out of 4 in 1p18
Go back to
Magnesium binding site 2 out
of 4 in the Hypoxanthine Phosphoribosyltransferase From Trypanosoma Cruzi, K68R Mutant, Ternary Substrates Complex
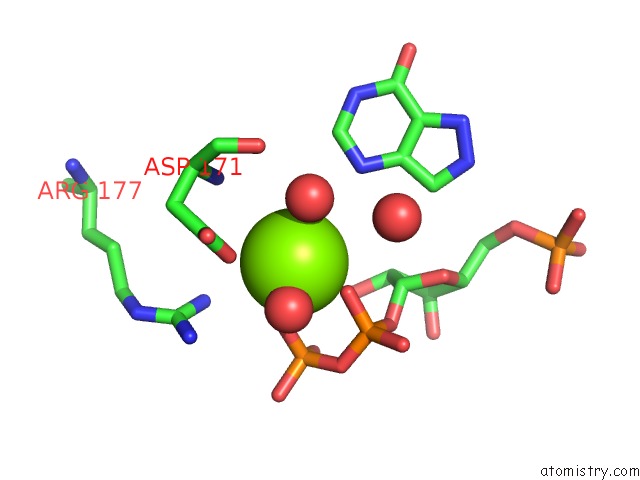
Mono view
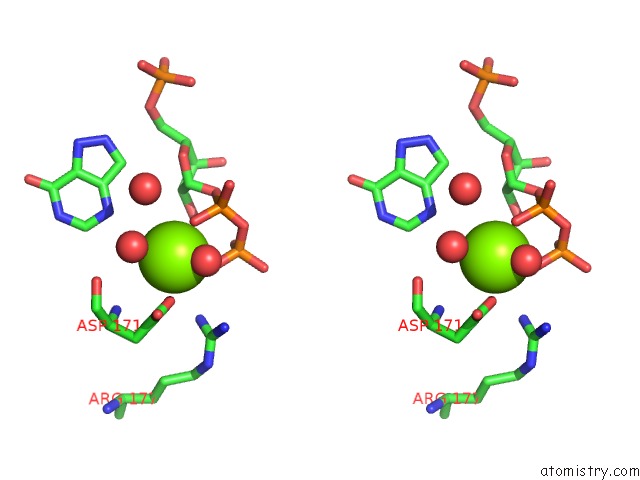
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Hypoxanthine Phosphoribosyltransferase From Trypanosoma Cruzi, K68R Mutant, Ternary Substrates Complex within 5.0Å range:
|
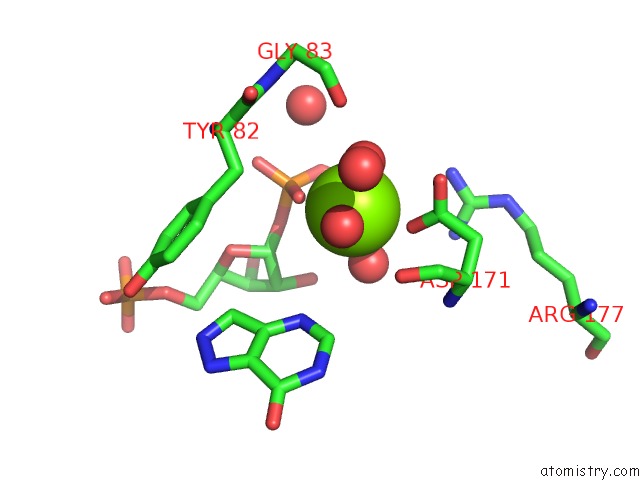
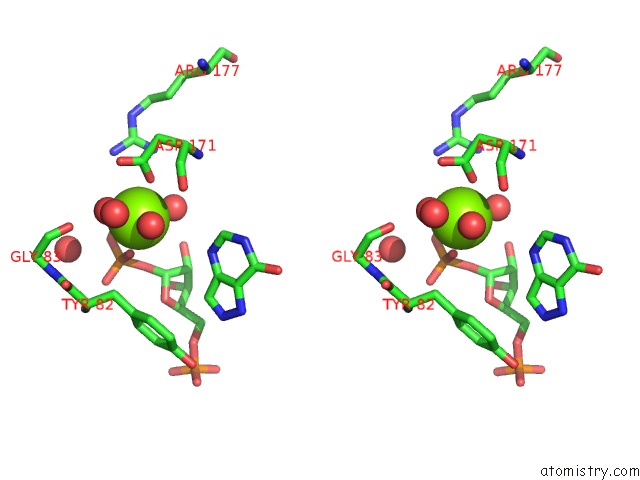
Magnesium binding site 3 out of 4 in 1p18
Go back to
Magnesium binding site 3 out
of 4 in the Hypoxanthine Phosphoribosyltransferase From Trypanosoma Cruzi, K68R Mutant, Ternary Substrates Complex
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 3 of Hypoxanthine Phosphoribosyltransferase From Trypanosoma Cruzi, K68R Mutant, Ternary Substrates Complex within 5.0Å range:
|
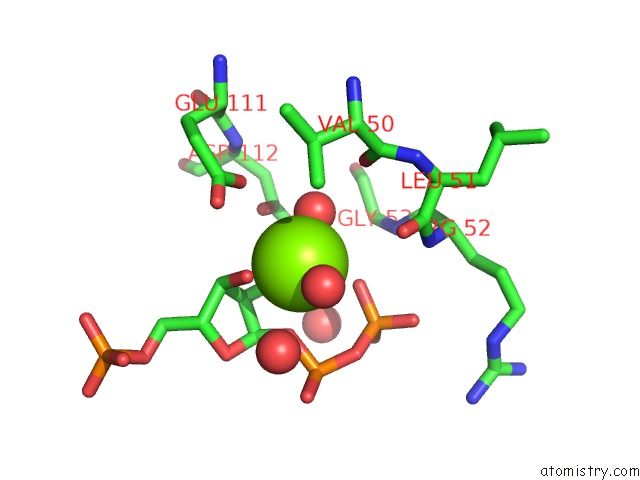
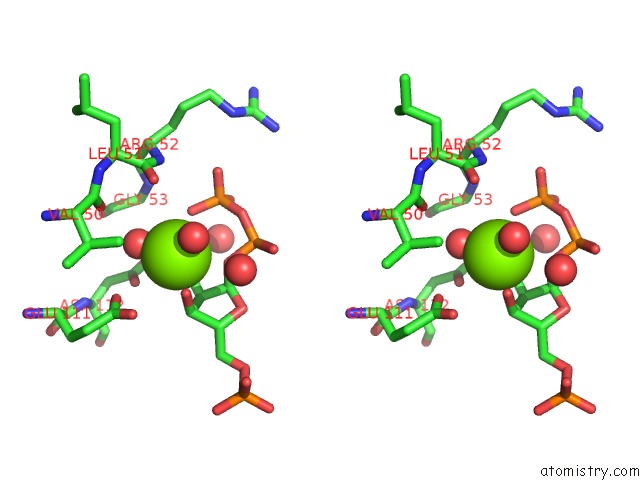
Magnesium binding site 4 out of 4 in 1p18
Go back to
Magnesium binding site 4 out
of 4 in the Hypoxanthine Phosphoribosyltransferase From Trypanosoma Cruzi, K68R Mutant, Ternary Substrates Complex
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 4 of Hypoxanthine Phosphoribosyltransferase From Trypanosoma Cruzi, K68R Mutant, Ternary Substrates Complex within 5.0Å range:
|
Reference:
B.Canyuk,
F.J.Medrano,
M.A.Wenck,
P.J.Focia,
A.E.Eakin,
S.P.Craig Iii.
Interactions at the Dimer Interface Influence the Relative Efficiencies For Purine Nucleotide Synthesis and Pyrophosphorolysis in A Phosphoribosyltransferase J.Mol.Biol. V. 335 905 2004.
ISSN: ISSN 0022-2836
PubMed: 14698288
DOI: 10.1016/J.JMB.2003.11.012
Page generated: Tue Aug 13 10:46:13 2024
ISSN: ISSN 0022-2836
PubMed: 14698288
DOI: 10.1016/J.JMB.2003.11.012
Last articles
Fe in 2AT0Fe in 2AUV
Fe in 2AUR
Fe in 2AUQ
Fe in 2AUP
Fe in 2AUO
Fe in 2ATJ
Fe in 2AT6
Fe in 2AT5
Fe in 2AT8