Magnesium »
PDB 1rtz-1s8f »
1s16 »
Magnesium in PDB 1s16: Crystal Structure of E. Coli Topoisomerase IV Pare 43KDA Subunit Complexed with Adpnp
Protein crystallography data
The structure of Crystal Structure of E. Coli Topoisomerase IV Pare 43KDA Subunit Complexed with Adpnp, PDB code: 1s16
was solved by
Y.Wei,
C.H.Gross,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 20.00 / 2.10 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 94.900, 120.800, 136.800, 90.00, 90.00, 90.00 |
| R / Rfree (%) | n/a / n/a |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Crystal Structure of E. Coli Topoisomerase IV Pare 43KDA Subunit Complexed with Adpnp
(pdb code 1s16). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 2 binding sites of Magnesium where determined in the Crystal Structure of E. Coli Topoisomerase IV Pare 43KDA Subunit Complexed with Adpnp, PDB code: 1s16:
Jump to Magnesium binding site number: 1; 2;
In total 2 binding sites of Magnesium where determined in the Crystal Structure of E. Coli Topoisomerase IV Pare 43KDA Subunit Complexed with Adpnp, PDB code: 1s16:
Jump to Magnesium binding site number: 1; 2;
Magnesium binding site 1 out of 2 in 1s16
Go back to
Magnesium binding site 1 out
of 2 in the Crystal Structure of E. Coli Topoisomerase IV Pare 43KDA Subunit Complexed with Adpnp
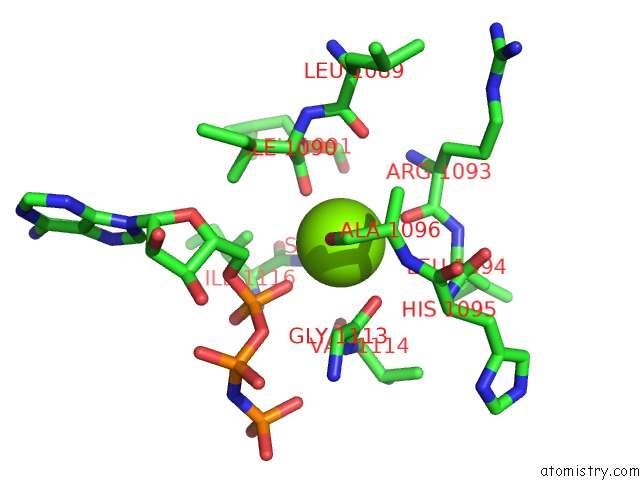
Mono view
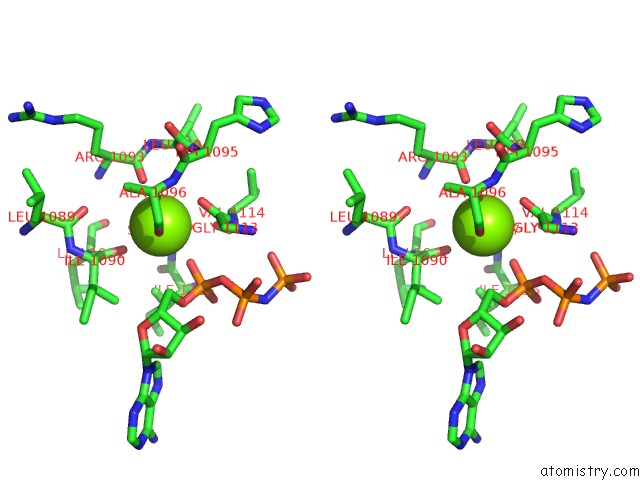
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Crystal Structure of E. Coli Topoisomerase IV Pare 43KDA Subunit Complexed with Adpnp within 5.0Å range:
|
Magnesium binding site 2 out of 2 in 1s16
Go back to
Magnesium binding site 2 out
of 2 in the Crystal Structure of E. Coli Topoisomerase IV Pare 43KDA Subunit Complexed with Adpnp
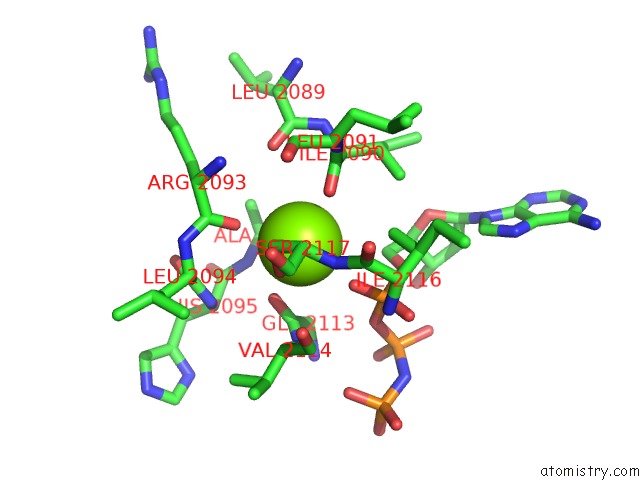
Mono view
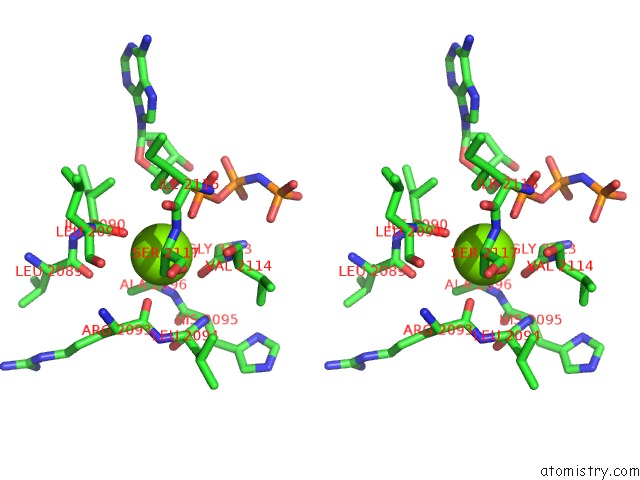
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Crystal Structure of E. Coli Topoisomerase IV Pare 43KDA Subunit Complexed with Adpnp within 5.0Å range:
|
Reference:
S.Bellon,
J.D.Parsons,
Y.Wei,
K.Hayakawa,
L.L.Swenson,
P.S.Charifson,
J.A.Lippke,
R.Aldape,
C.H.Gross.
Crystal Structures of Escherichia Coli Topoisomerase IV Pare Subunit (24 and 43 Kilodaltons): A Single Residue Dictates Differences in Novobiocin Potency Against Topoisomerase IV and Dna Gyrase. Antimicrob.Agents Chemother. V. 48 1856 2004.
ISSN: ISSN 0066-4804
PubMed: 15105144
DOI: 10.1128/AAC.48.5.1856-1864.2004
Page generated: Sun Aug 10 04:01:35 2025
ISSN: ISSN 0066-4804
PubMed: 15105144
DOI: 10.1128/AAC.48.5.1856-1864.2004
Last articles
Mg in 6HW5Mg in 6HW6
Mg in 6HW4
Mg in 6HVY
Mg in 6HW3
Mg in 6HW0
Mg in 6HW1
Mg in 6HVX
Mg in 6HQB
Mg in 6HVW