Magnesium »
PDB 1vq5-1w55 »
1w49 »
Magnesium in PDB 1w49: P4 Protein From Bacteriophage PHI12 in Complex with Ampcpp and Mg
Protein crystallography data
The structure of P4 Protein From Bacteriophage PHI12 in Complex with Ampcpp and Mg, PDB code: 1w49
was solved by
E.J.Mancini,
D.E.Kainov,
J.M.Grimes,
R.Tuma,
D.H.Bamford,
D.I.Stuart,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 30.00 / 2.40 |
| Space group | I 2 2 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 105.700, 130.100, 159.200, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 18.8 / 24.6 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the P4 Protein From Bacteriophage PHI12 in Complex with Ampcpp and Mg
(pdb code 1w49). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 3 binding sites of Magnesium where determined in the P4 Protein From Bacteriophage PHI12 in Complex with Ampcpp and Mg, PDB code: 1w49:
Jump to Magnesium binding site number: 1; 2; 3;
In total 3 binding sites of Magnesium where determined in the P4 Protein From Bacteriophage PHI12 in Complex with Ampcpp and Mg, PDB code: 1w49:
Jump to Magnesium binding site number: 1; 2; 3;
Magnesium binding site 1 out of 3 in 1w49
Go back to
Magnesium binding site 1 out
of 3 in the P4 Protein From Bacteriophage PHI12 in Complex with Ampcpp and Mg
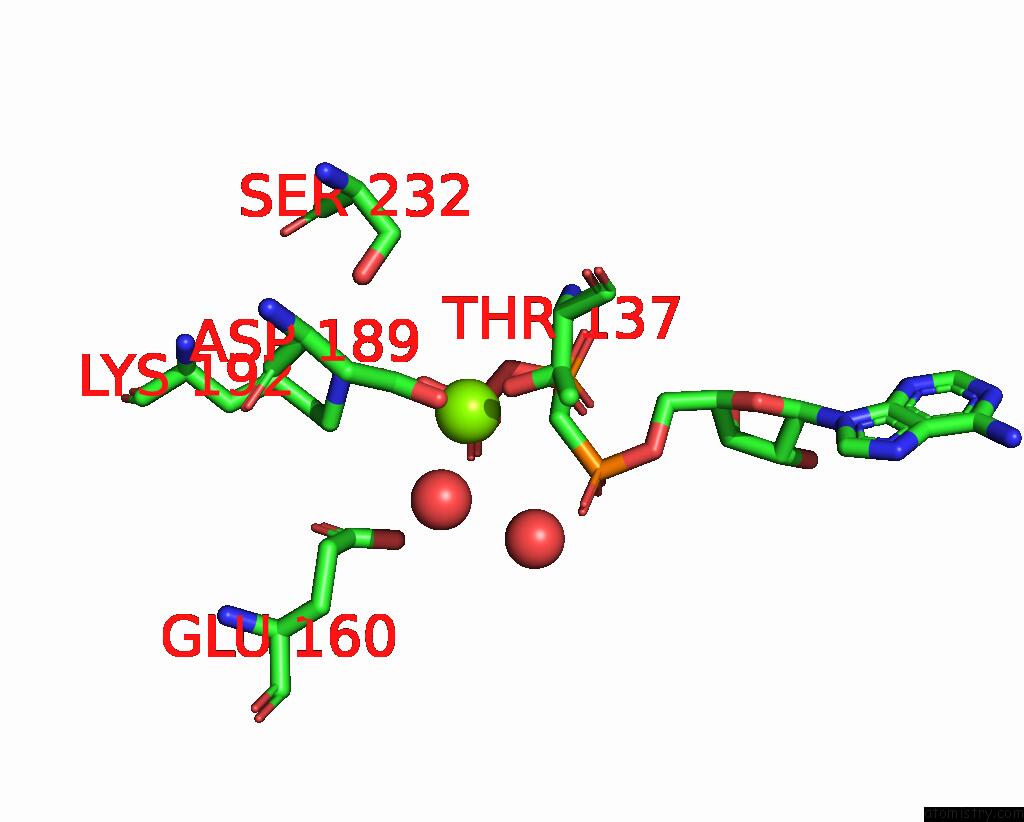
Mono view
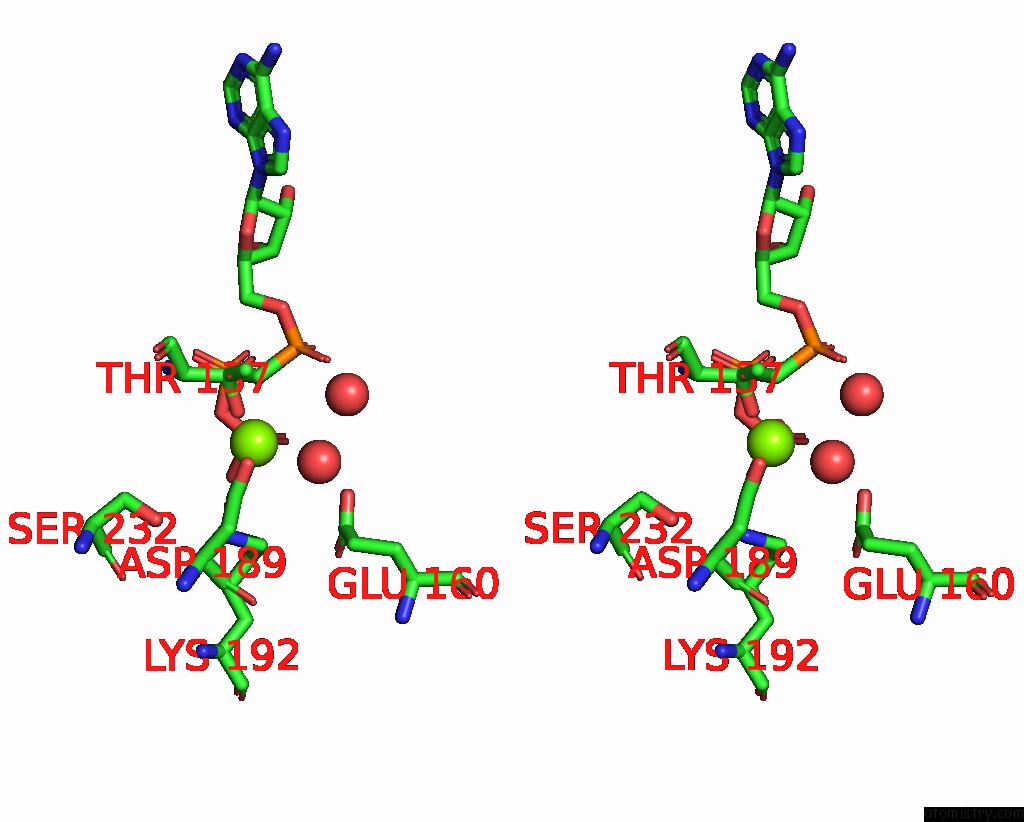
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of P4 Protein From Bacteriophage PHI12 in Complex with Ampcpp and Mg within 5.0Å range:
|
Magnesium binding site 2 out of 3 in 1w49
Go back to
Magnesium binding site 2 out
of 3 in the P4 Protein From Bacteriophage PHI12 in Complex with Ampcpp and Mg
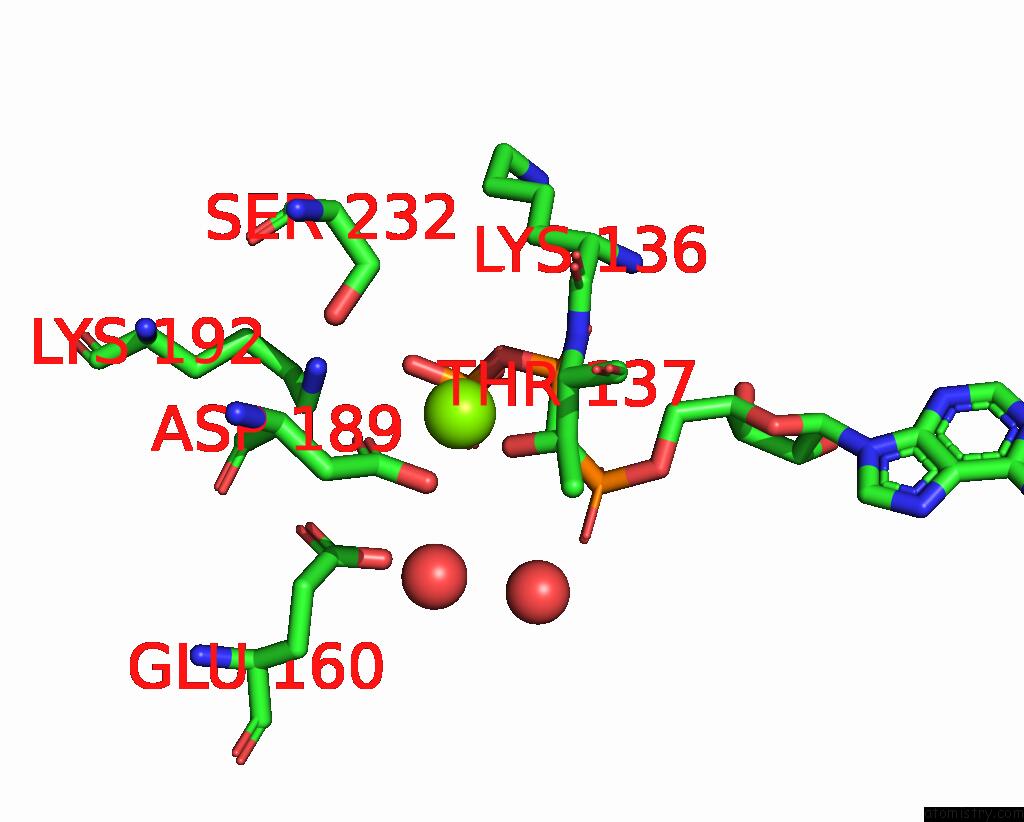
Mono view
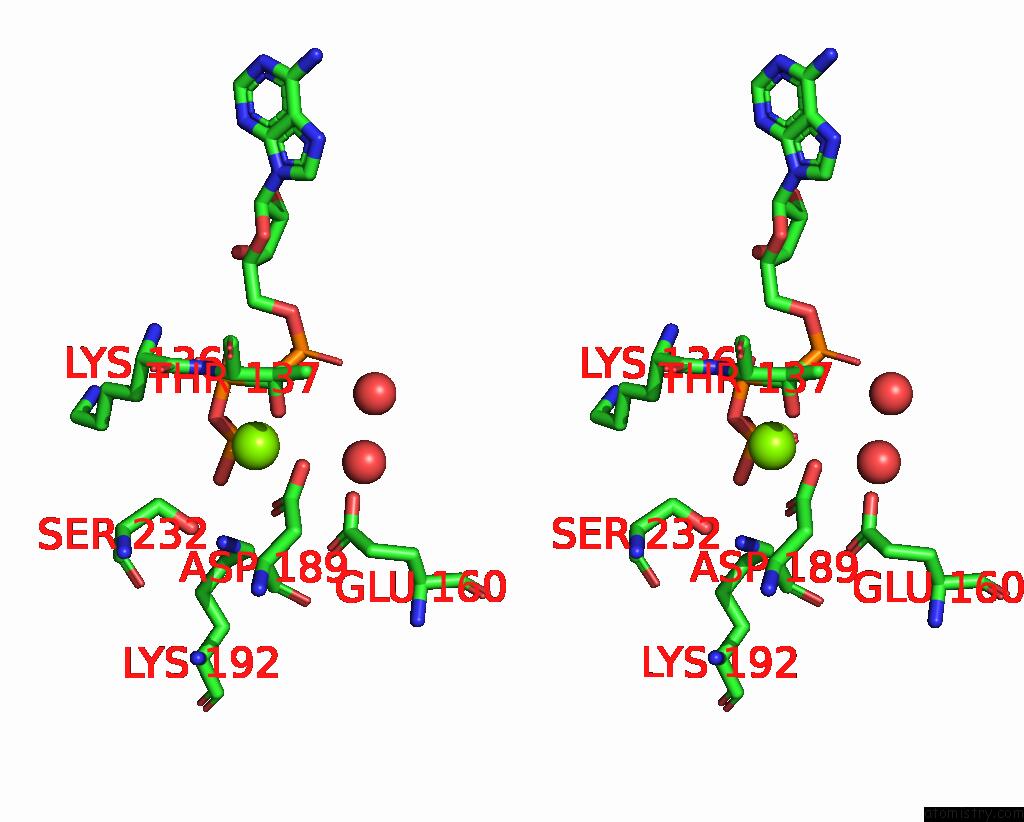
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of P4 Protein From Bacteriophage PHI12 in Complex with Ampcpp and Mg within 5.0Å range:
|
Magnesium binding site 3 out of 3 in 1w49
Go back to
Magnesium binding site 3 out
of 3 in the P4 Protein From Bacteriophage PHI12 in Complex with Ampcpp and Mg
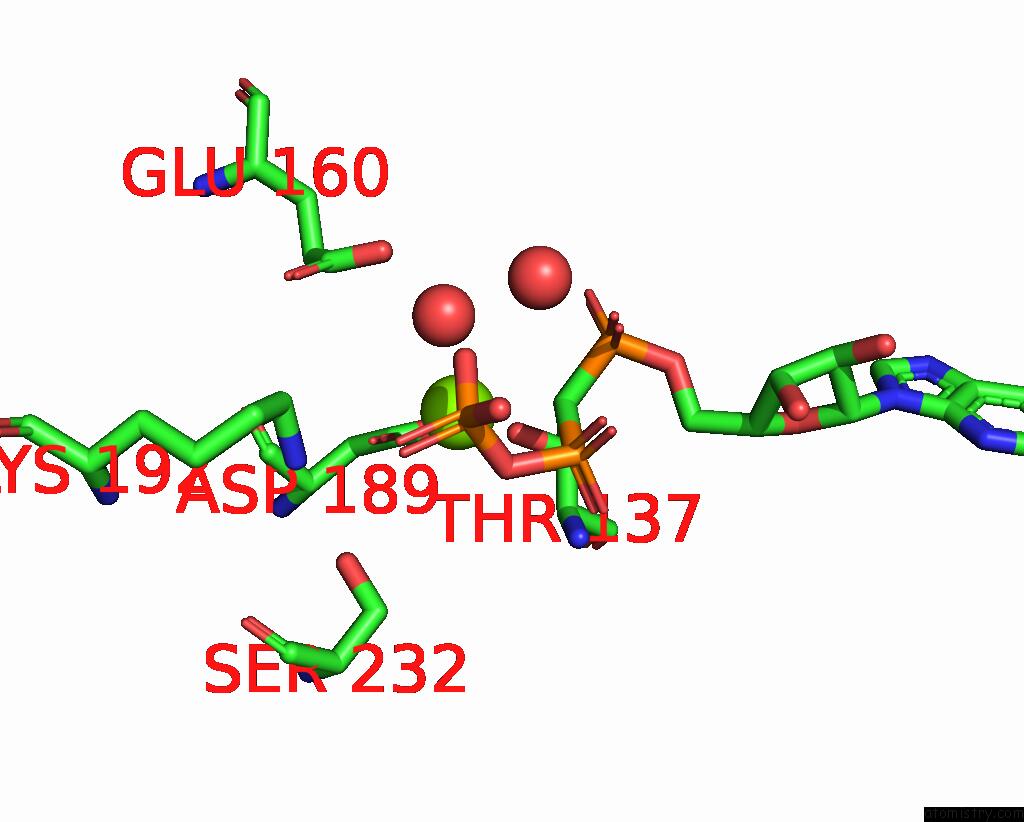
Mono view
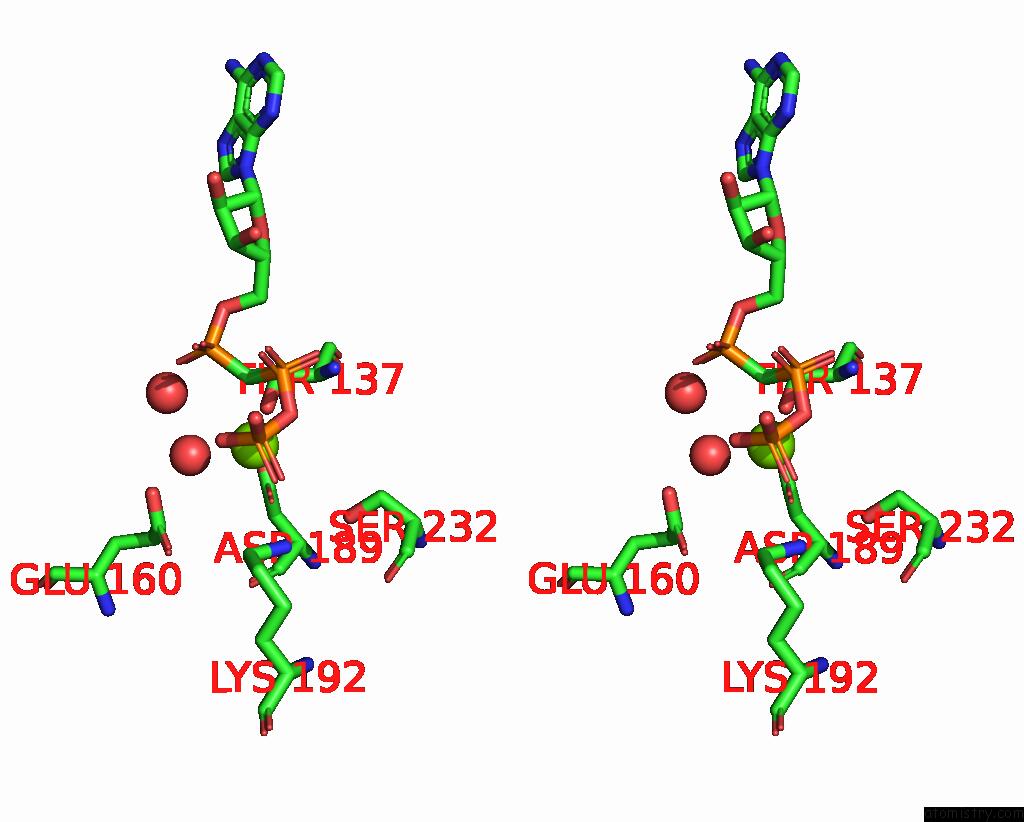
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 3 of P4 Protein From Bacteriophage PHI12 in Complex with Ampcpp and Mg within 5.0Å range:
|
Reference:
E.J.Mancini,
D.E.Kainov,
J.M.Grimes,
R.Tuma,
D.H.Bamford,
D.I.Stuart.
Atomic Snapshots of An Rna Packaging Motor Reveal Conformational Changes Linking Atp Hydrolysis to Rna Translocation Cell(Cambridge,Mass.) V. 118 743 2004.
ISSN: ISSN 0092-8674
PubMed: 15369673
DOI: 10.1016/J.CELL.2004.09.007
Page generated: Sun Aug 10 06:33:15 2025
ISSN: ISSN 0092-8674
PubMed: 15369673
DOI: 10.1016/J.CELL.2004.09.007
Last articles
Mg in 2PS5Mg in 2PS7
Mg in 2PS2
Mg in 2PS4
Mg in 2PRC
Mg in 2PS1
Mg in 2PRY
Mg in 2PRN
Mg in 2PPQ
Mg in 2PPB