Magnesium »
PDB 2amc-2b2k »
2b1q »
Magnesium in PDB 2b1q: X-Ray Structure of the Sucrose-Phosphatase (Spp) From Synechocystis Sp.PCC6803 in Complex with Trehalose
Enzymatic activity of X-Ray Structure of the Sucrose-Phosphatase (Spp) From Synechocystis Sp.PCC6803 in Complex with Trehalose
All present enzymatic activity of X-Ray Structure of the Sucrose-Phosphatase (Spp) From Synechocystis Sp.PCC6803 in Complex with Trehalose:
3.1.3.24;
3.1.3.24;
Protein crystallography data
The structure of X-Ray Structure of the Sucrose-Phosphatase (Spp) From Synechocystis Sp.PCC6803 in Complex with Trehalose, PDB code: 2b1q
was solved by
S.Fieulaine,
J.E.Lunn,
J.-L.Ferrer,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 50.00 / 2.20 |
| Space group | P 65 2 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 68.910, 68.910, 268.650, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 17.8 / 20 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the X-Ray Structure of the Sucrose-Phosphatase (Spp) From Synechocystis Sp.PCC6803 in Complex with Trehalose
(pdb code 2b1q). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total only one binding site of Magnesium was determined in the X-Ray Structure of the Sucrose-Phosphatase (Spp) From Synechocystis Sp.PCC6803 in Complex with Trehalose, PDB code: 2b1q:
In total only one binding site of Magnesium was determined in the X-Ray Structure of the Sucrose-Phosphatase (Spp) From Synechocystis Sp.PCC6803 in Complex with Trehalose, PDB code: 2b1q:
Magnesium binding site 1 out of 1 in 2b1q
Go back to
Magnesium binding site 1 out
of 1 in the X-Ray Structure of the Sucrose-Phosphatase (Spp) From Synechocystis Sp.PCC6803 in Complex with Trehalose
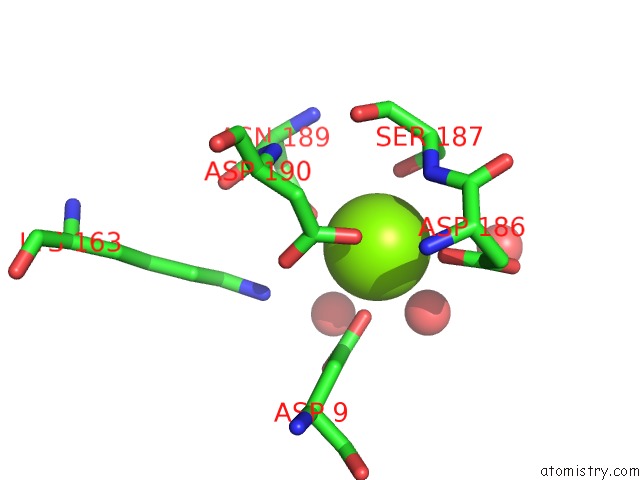
Mono view
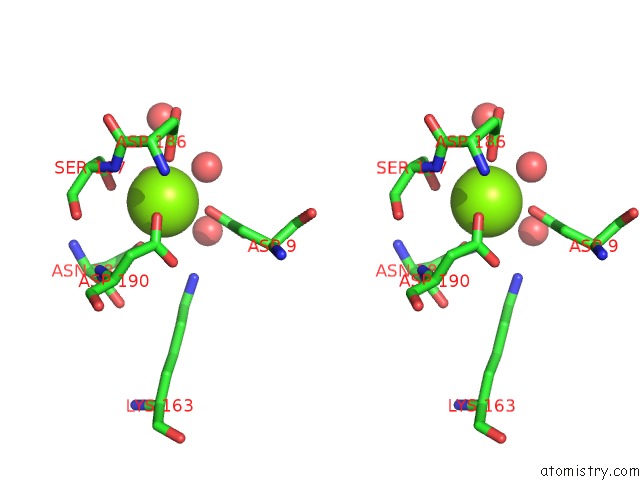
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of X-Ray Structure of the Sucrose-Phosphatase (Spp) From Synechocystis Sp.PCC6803 in Complex with Trehalose within 5.0Å range:
|
Reference:
S.Fieulaine,
J.E.Lunn,
J.-L.Ferrer.
Crystal Structure of A Cyanobacterial Sucrose-Phosphatase in Complex with Glucose-Containing Disaccharides Proteins V. 68 796 2007.
ISSN: ISSN 0887-3585
PubMed: 17510968
DOI: 10.1002/PROT.21481
Page generated: Sun Aug 10 09:52:12 2025
ISSN: ISSN 0887-3585
PubMed: 17510968
DOI: 10.1002/PROT.21481
Last articles
Mg in 2WSBMg in 2WPD
Mg in 2WOQ
Mg in 2WOJ
Mg in 2WQS
Mg in 2WQN
Mg in 2WOG
Mg in 2WNL
Mg in 2WLL
Mg in 2WNH