Magnesium »
PDB 2cw0-2dkg »
2d5h »
Magnesium in PDB 2d5h: Crystal Structure of Recombinant Soybean Proglycinin A3B4 Subunit, Its Comparison with Mature Glycinin A3B4 Subunit, Responsible For Hexamer Assembly
Protein crystallography data
The structure of Crystal Structure of Recombinant Soybean Proglycinin A3B4 Subunit, Its Comparison with Mature Glycinin A3B4 Subunit, Responsible For Hexamer Assembly, PDB code: 2d5h
was solved by
T.Itoh,
M.Adachi,
T.Masuda,
B.Mikami,
S.Utsumi,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 15.00 / 2.80 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 89.378, 223.738, 88.660, 90.00, 119.85, 90.00 |
| R / Rfree (%) | 24.8 / 32.5 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Crystal Structure of Recombinant Soybean Proglycinin A3B4 Subunit, Its Comparison with Mature Glycinin A3B4 Subunit, Responsible For Hexamer Assembly
(pdb code 2d5h). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 6 binding sites of Magnesium where determined in the Crystal Structure of Recombinant Soybean Proglycinin A3B4 Subunit, Its Comparison with Mature Glycinin A3B4 Subunit, Responsible For Hexamer Assembly, PDB code: 2d5h:
Jump to Magnesium binding site number: 1; 2; 3; 4; 5; 6;
In total 6 binding sites of Magnesium where determined in the Crystal Structure of Recombinant Soybean Proglycinin A3B4 Subunit, Its Comparison with Mature Glycinin A3B4 Subunit, Responsible For Hexamer Assembly, PDB code: 2d5h:
Jump to Magnesium binding site number: 1; 2; 3; 4; 5; 6;
Magnesium binding site 1 out of 6 in 2d5h
Go back to
Magnesium binding site 1 out
of 6 in the Crystal Structure of Recombinant Soybean Proglycinin A3B4 Subunit, Its Comparison with Mature Glycinin A3B4 Subunit, Responsible For Hexamer Assembly

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Crystal Structure of Recombinant Soybean Proglycinin A3B4 Subunit, Its Comparison with Mature Glycinin A3B4 Subunit, Responsible For Hexamer Assembly within 5.0Å range:
|
Magnesium binding site 2 out of 6 in 2d5h
Go back to
Magnesium binding site 2 out
of 6 in the Crystal Structure of Recombinant Soybean Proglycinin A3B4 Subunit, Its Comparison with Mature Glycinin A3B4 Subunit, Responsible For Hexamer Assembly

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Crystal Structure of Recombinant Soybean Proglycinin A3B4 Subunit, Its Comparison with Mature Glycinin A3B4 Subunit, Responsible For Hexamer Assembly within 5.0Å range:
|
Magnesium binding site 3 out of 6 in 2d5h
Go back to
Magnesium binding site 3 out
of 6 in the Crystal Structure of Recombinant Soybean Proglycinin A3B4 Subunit, Its Comparison with Mature Glycinin A3B4 Subunit, Responsible For Hexamer Assembly

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 3 of Crystal Structure of Recombinant Soybean Proglycinin A3B4 Subunit, Its Comparison with Mature Glycinin A3B4 Subunit, Responsible For Hexamer Assembly within 5.0Å range:
|
Magnesium binding site 4 out of 6 in 2d5h
Go back to
Magnesium binding site 4 out
of 6 in the Crystal Structure of Recombinant Soybean Proglycinin A3B4 Subunit, Its Comparison with Mature Glycinin A3B4 Subunit, Responsible For Hexamer Assembly

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 4 of Crystal Structure of Recombinant Soybean Proglycinin A3B4 Subunit, Its Comparison with Mature Glycinin A3B4 Subunit, Responsible For Hexamer Assembly within 5.0Å range:
|
Magnesium binding site 5 out of 6 in 2d5h
Go back to
Magnesium binding site 5 out
of 6 in the Crystal Structure of Recombinant Soybean Proglycinin A3B4 Subunit, Its Comparison with Mature Glycinin A3B4 Subunit, Responsible For Hexamer Assembly

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 5 of Crystal Structure of Recombinant Soybean Proglycinin A3B4 Subunit, Its Comparison with Mature Glycinin A3B4 Subunit, Responsible For Hexamer Assembly within 5.0Å range:
|
Magnesium binding site 6 out of 6 in 2d5h
Go back to
Magnesium binding site 6 out
of 6 in the Crystal Structure of Recombinant Soybean Proglycinin A3B4 Subunit, Its Comparison with Mature Glycinin A3B4 Subunit, Responsible For Hexamer Assembly
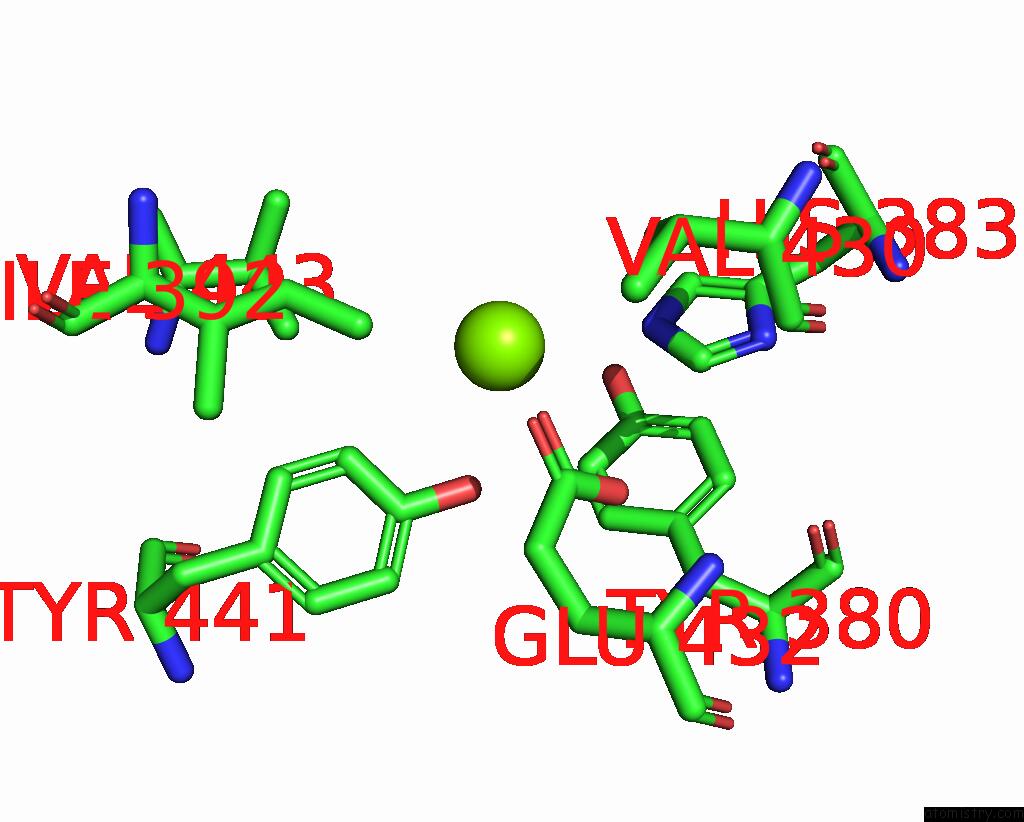
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 6 of Crystal Structure of Recombinant Soybean Proglycinin A3B4 Subunit, Its Comparison with Mature Glycinin A3B4 Subunit, Responsible For Hexamer Assembly within 5.0Å range:
|
Reference:
M.R.Tandang-Silvas,
T.Fukuda,
C.Fukuda,
K.Prak,
C.Cabanos,
A.Kimura,
T.Itoh,
B.Mikami,
S.Utsumi,
N.Maruyama.
Conservation and Divergence on Plant Seed 11S Globulins Based on Crystal Structures. Biochim.Biophys.Acta 2010.
ISSN: ISSN 0006-3002
PubMed: 20215054
DOI: 10.1016/J.BBAPAP.2010.02.016
Page generated: Tue Aug 13 22:25:43 2024
ISSN: ISSN 0006-3002
PubMed: 20215054
DOI: 10.1016/J.BBAPAP.2010.02.016
Last articles
Ca in 5OSTCa in 5OSK
Ca in 5OLR
Ca in 5OLW
Ca in 5OP3
Ca in 5ONP
Ca in 5ONQ
Ca in 5OLS
Ca in 5OLP
Ca in 5OLQ