Magnesium »
PDB 2dlc-2e74 »
2dua »
Magnesium in PDB 2dua: Crystal Structure of Phosphonopyruvate Hydrolase Complex with Oxalate and Mg++
Protein crystallography data
The structure of Crystal Structure of Phosphonopyruvate Hydrolase Complex with Oxalate and Mg++, PDB code: 2dua
was solved by
O.Herzberg,
C.Chen,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 32.78 / 2.00 |
| Space group | I 2 2 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 74.890, 79.180, 93.430, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 15.6 / 21.9 |
Other elements in 2dua:
The structure of Crystal Structure of Phosphonopyruvate Hydrolase Complex with Oxalate and Mg++ also contains other interesting chemical elements:
| Chlorine | (Cl) | 2 atoms |
| Sodium | (Na) | 1 atom |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Crystal Structure of Phosphonopyruvate Hydrolase Complex with Oxalate and Mg++
(pdb code 2dua). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total only one binding site of Magnesium was determined in the Crystal Structure of Phosphonopyruvate Hydrolase Complex with Oxalate and Mg++, PDB code: 2dua:
In total only one binding site of Magnesium was determined in the Crystal Structure of Phosphonopyruvate Hydrolase Complex with Oxalate and Mg++, PDB code: 2dua:
Magnesium binding site 1 out of 1 in 2dua
Go back to
Magnesium binding site 1 out
of 1 in the Crystal Structure of Phosphonopyruvate Hydrolase Complex with Oxalate and Mg++
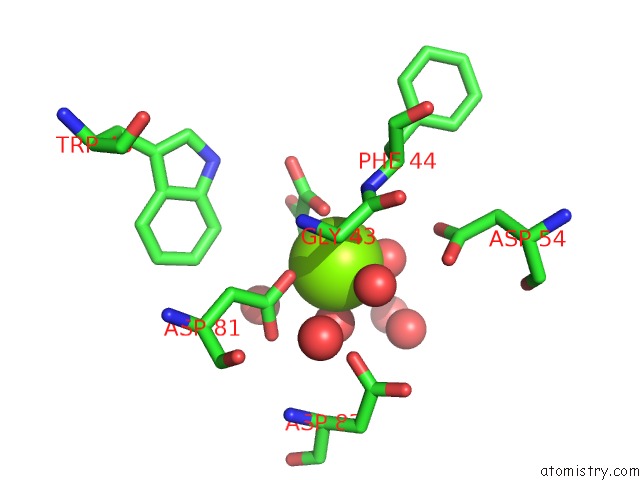
Mono view
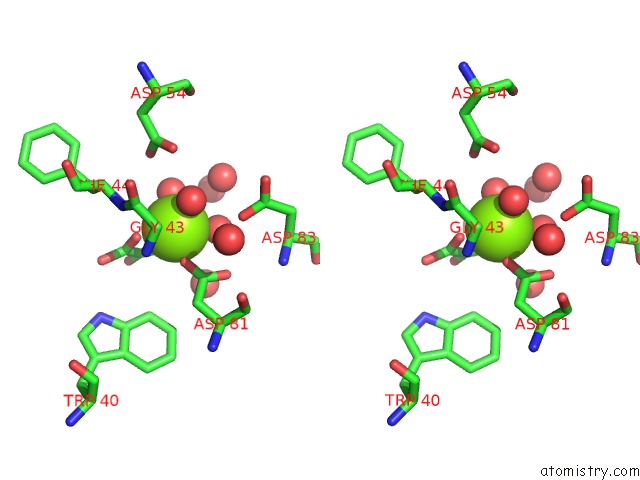
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Crystal Structure of Phosphonopyruvate Hydrolase Complex with Oxalate and Mg++ within 5.0Å range:
|
Reference:
C.C.H.Chen,
Y.Han,
W.Niu,
A.N.Kulakova,
A.Howard,
J.P.Quinn,
D.Dunaway-Mariano,
O.Herzberg.
Structure and Kinetics of Phosphonopyruvate Hydrolase From Voriovorax Sp. PAL2: New Insight Into the Divergence of Catalysis Within the Pep Mutase/Isocitrate Lyase Superfamily Biochemistry V. 45 11491 2006.
ISSN: ISSN 0006-2960
PubMed: 16981709
DOI: 10.1021/BI061208L
Page generated: Tue Aug 13 22:35:53 2024
ISSN: ISSN 0006-2960
PubMed: 16981709
DOI: 10.1021/BI061208L
Last articles
Ca in 5SZLCa in 5SY1
Ca in 5SWI
Ca in 5SVE
Ca in 5SSX
Ca in 5SV0
Ca in 5STD
Ca in 5SSZ
Ca in 5SSY
Ca in 5SIC