Magnesium »
PDB 2e75-2ein »
2e8a »
Magnesium in PDB 2e8a: Crystal Structure of the Human HSP70 Atpase Domain in Complex with Amp-Pnp
Protein crystallography data
The structure of Crystal Structure of the Human HSP70 Atpase Domain in Complex with Amp-Pnp, PDB code: 2e8a
was solved by
M.Shida,
R.Ishii,
T.Takagi,
S.Kishishita,
M.Shirouzu,
S.Yokoyama,
Riken Structural Genomics/Proteomics Initiative (Rsgi),
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 46.94 / 1.77 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 45.490, 62.220, 143.040, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 18.5 / 23 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Crystal Structure of the Human HSP70 Atpase Domain in Complex with Amp-Pnp
(pdb code 2e8a). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total only one binding site of Magnesium was determined in the Crystal Structure of the Human HSP70 Atpase Domain in Complex with Amp-Pnp, PDB code: 2e8a:
In total only one binding site of Magnesium was determined in the Crystal Structure of the Human HSP70 Atpase Domain in Complex with Amp-Pnp, PDB code: 2e8a:
Magnesium binding site 1 out of 1 in 2e8a
Go back to
Magnesium binding site 1 out
of 1 in the Crystal Structure of the Human HSP70 Atpase Domain in Complex with Amp-Pnp
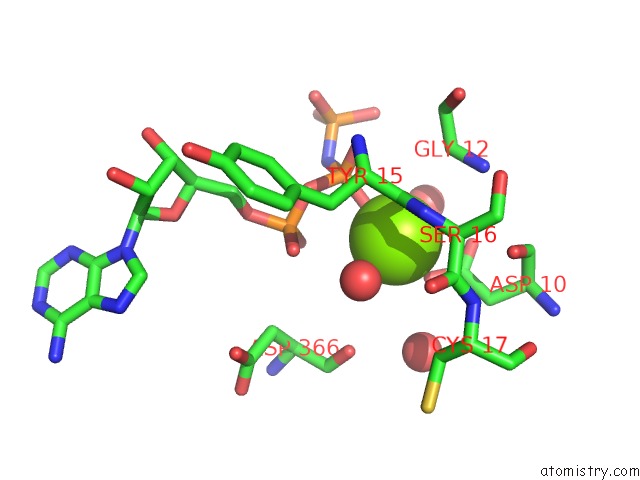
Mono view
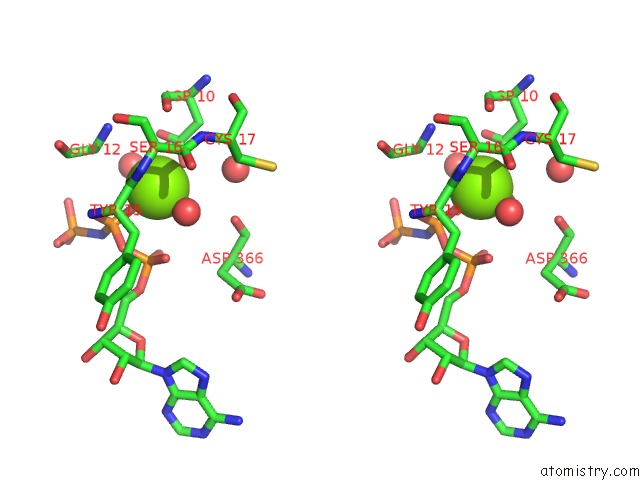
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Crystal Structure of the Human HSP70 Atpase Domain in Complex with Amp-Pnp within 5.0Å range:
|
Reference:
M.Shida,
A.Arakawa,
R.Ishii,
S.Kishishita,
T.Takagi,
M.Kukimoto-Niino,
S.Sugano,
A.Tanaka,
M.Shirouzu,
S.Yokoyama.
Direct Inter-Subdomain Interactions Switch Between the Closed and Open Forms of the HSP70 Nucleotide-Binding Domain in the Nucleotide-Free State. Acta Crystallogr.,Sect.D V. 66 223 2010.
ISSN: ISSN 0907-4449
PubMed: 20179333
DOI: 10.1107/S0907444909053979
Page generated: Tue Aug 13 22:41:22 2024
ISSN: ISSN 0907-4449
PubMed: 20179333
DOI: 10.1107/S0907444909053979
Last articles
Cl in 5W4RCl in 5W4P
Cl in 5W4O
Cl in 5W16
Cl in 5W15
Cl in 5W4M
Cl in 5W4I
Cl in 5W4L
Cl in 5W44
Cl in 5W4D