Magnesium »
PDB 2fx3-2g9y »
2g28 »
Magnesium in PDB 2g28: E. Coli Pyruvate Dehydrogenase H407A Variant Phosphonolactylthiamin Diphosphate Complex
Enzymatic activity of E. Coli Pyruvate Dehydrogenase H407A Variant Phosphonolactylthiamin Diphosphate Complex
All present enzymatic activity of E. Coli Pyruvate Dehydrogenase H407A Variant Phosphonolactylthiamin Diphosphate Complex:
1.2.4.1;
1.2.4.1;
Protein crystallography data
The structure of E. Coli Pyruvate Dehydrogenase H407A Variant Phosphonolactylthiamin Diphosphate Complex, PDB code: 2g28
was solved by
W.Furey,
P.Arjunan,
K.Chandrasekhar,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 41.77 / 1.85 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 81.990, 143.200, 82.530, 90.00, 102.61, 90.00 |
| R / Rfree (%) | 21.6 / 24 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the E. Coli Pyruvate Dehydrogenase H407A Variant Phosphonolactylthiamin Diphosphate Complex
(pdb code 2g28). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 2 binding sites of Magnesium where determined in the E. Coli Pyruvate Dehydrogenase H407A Variant Phosphonolactylthiamin Diphosphate Complex, PDB code: 2g28:
Jump to Magnesium binding site number: 1; 2;
In total 2 binding sites of Magnesium where determined in the E. Coli Pyruvate Dehydrogenase H407A Variant Phosphonolactylthiamin Diphosphate Complex, PDB code: 2g28:
Jump to Magnesium binding site number: 1; 2;
Magnesium binding site 1 out of 2 in 2g28
Go back to
Magnesium binding site 1 out
of 2 in the E. Coli Pyruvate Dehydrogenase H407A Variant Phosphonolactylthiamin Diphosphate Complex
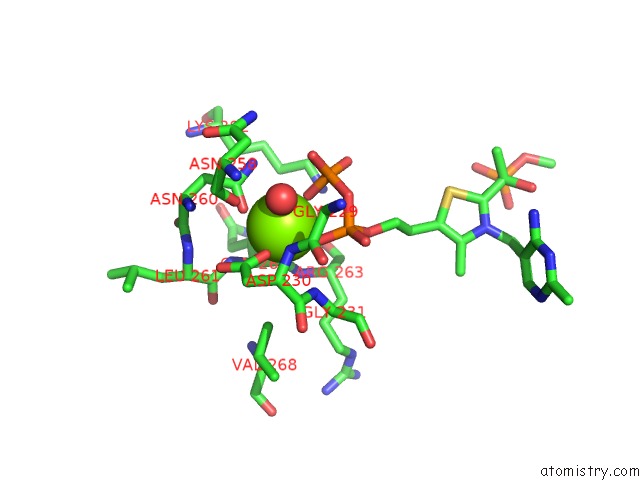
Mono view
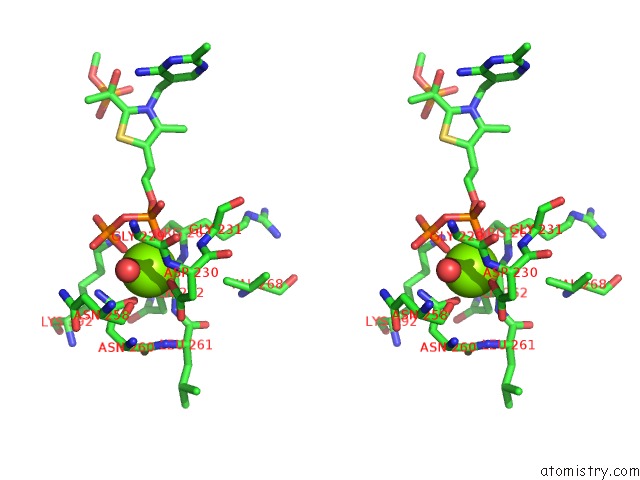
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of E. Coli Pyruvate Dehydrogenase H407A Variant Phosphonolactylthiamin Diphosphate Complex within 5.0Å range:
|
Magnesium binding site 2 out of 2 in 2g28
Go back to
Magnesium binding site 2 out
of 2 in the E. Coli Pyruvate Dehydrogenase H407A Variant Phosphonolactylthiamin Diphosphate Complex
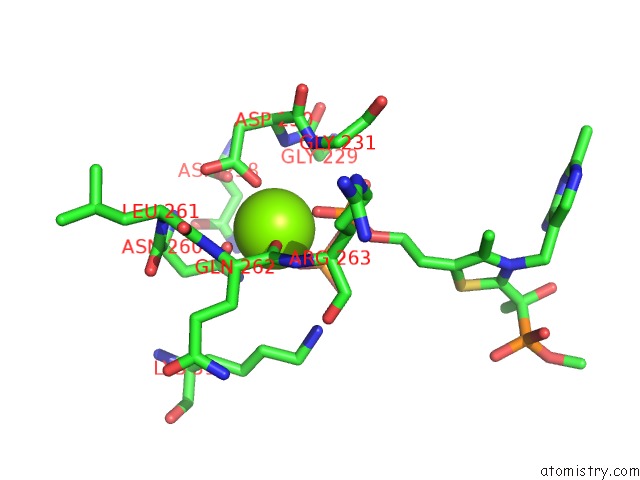
Mono view
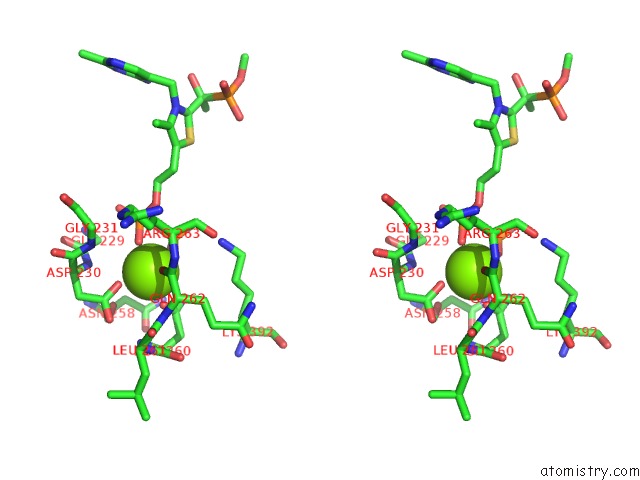
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of E. Coli Pyruvate Dehydrogenase H407A Variant Phosphonolactylthiamin Diphosphate Complex within 5.0Å range:
|
Reference:
P.Arjunan,
M.Sax,
A.Brunskill,
K.Chandrasekhar,
N.Nemeria,
S.Zhang,
F.Jordan,
W.Furey.
A Thiamin-Bound, Pre-Decarboxylation Reaction Intermediate Analogue in the Pyruvate Dehydrogenase E1 Subunit Induces Large Scale Disorder-to-Order Transformations in the Enzyme and Reveals Novel Structural Features in the Covalently Bound Adduct. J.Biol.Chem. V. 281 15296 2006.
ISSN: ISSN 0021-9258
PubMed: 16531404
DOI: 10.1074/JBC.M600656200
Page generated: Tue Aug 13 23:22:11 2024
ISSN: ISSN 0021-9258
PubMed: 16531404
DOI: 10.1074/JBC.M600656200
Last articles
Cl in 5YQHCl in 5YRO
Cl in 5YKY
Cl in 5YQA
Cl in 5YPL
Cl in 5YPK
Cl in 5YPJ
Cl in 5YPI
Cl in 5YPD
Cl in 5YKG