Magnesium »
PDB 2iec-2ivn »
2iof »
Magnesium in PDB 2iof: Crystal Structure of Phosphonoacetaldehyde Hydrolase with Sodium Borohydride-Reduced Substrate Intermediate
Protein crystallography data
The structure of Crystal Structure of Phosphonoacetaldehyde Hydrolase with Sodium Borohydride-Reduced Substrate Intermediate, PDB code: 2iof
was solved by
K.A.Allen,
S.D.Lahiri,
G.Zhang,
D.Dunaway-Mariano,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 100.00 / 2.50 |
| Space group | C 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 159.820, 64.020, 98.330, 90.00, 123.52, 90.00 |
| R / Rfree (%) | 23.1 / 29.8 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Crystal Structure of Phosphonoacetaldehyde Hydrolase with Sodium Borohydride-Reduced Substrate Intermediate
(pdb code 2iof). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 2 binding sites of Magnesium where determined in the Crystal Structure of Phosphonoacetaldehyde Hydrolase with Sodium Borohydride-Reduced Substrate Intermediate, PDB code: 2iof:
Jump to Magnesium binding site number: 1; 2;
In total 2 binding sites of Magnesium where determined in the Crystal Structure of Phosphonoacetaldehyde Hydrolase with Sodium Borohydride-Reduced Substrate Intermediate, PDB code: 2iof:
Jump to Magnesium binding site number: 1; 2;
Magnesium binding site 1 out of 2 in 2iof
Go back to
Magnesium binding site 1 out
of 2 in the Crystal Structure of Phosphonoacetaldehyde Hydrolase with Sodium Borohydride-Reduced Substrate Intermediate
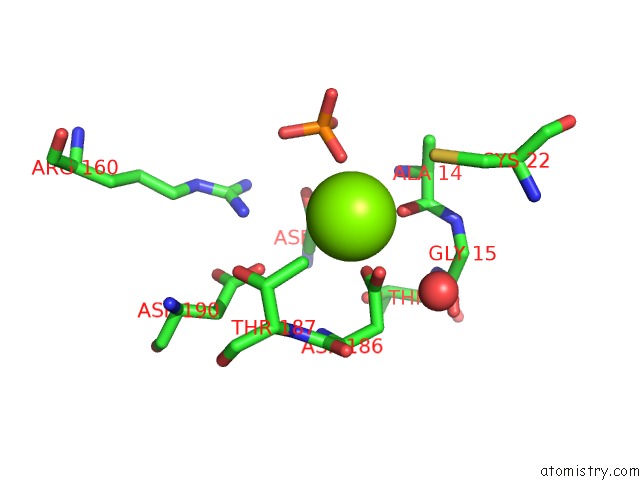
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Crystal Structure of Phosphonoacetaldehyde Hydrolase with Sodium Borohydride-Reduced Substrate Intermediate within 5.0Å range:
|
Magnesium binding site 2 out of 2 in 2iof
Go back to
Magnesium binding site 2 out
of 2 in the Crystal Structure of Phosphonoacetaldehyde Hydrolase with Sodium Borohydride-Reduced Substrate Intermediate

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Crystal Structure of Phosphonoacetaldehyde Hydrolase with Sodium Borohydride-Reduced Substrate Intermediate within 5.0Å range:
|
Reference:
S.D.Lahiri,
G.Zhang,
D.Dunaway-Mariano,
K.N.Allen.
Diversification of Function in the Haloacid Dehalogenase Enzyme Superfamily: the Role of the Cap Domain in Hydrolytic Phosphoruscarbon Bond Cleavage. Bioorg.Chem. V. 34 394 2006.
ISSN: ISSN 0045-2068
PubMed: 17070898
DOI: 10.1016/J.BIOORG.2006.09.007
Page generated: Wed Aug 14 00:13:47 2024
ISSN: ISSN 0045-2068
PubMed: 17070898
DOI: 10.1016/J.BIOORG.2006.09.007
Last articles
Ca in 5S5FCa in 5S5D
Ca in 5S5E
Ca in 5S5C
Ca in 5S5B
Ca in 5S5A
Ca in 5S59
Ca in 5S58
Ca in 5S57
Ca in 5S56