Magnesium »
PDB 2iec-2ivn »
2ior »
Magnesium in PDB 2ior: Crystal Structure of the N-Terminal Domain of Htpg, the Escherichia Coli HSP90, Bound to Adp
Protein crystallography data
The structure of Crystal Structure of the N-Terminal Domain of Htpg, the Escherichia Coli HSP90, Bound to Adp, PDB code: 2ior
was solved by
A.K.Shiau,
S.F.Harris,
D.A.Agard,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 55.90 / 1.65 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 38.494, 77.193, 80.479, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 16.3 / 19.8 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Crystal Structure of the N-Terminal Domain of Htpg, the Escherichia Coli HSP90, Bound to Adp
(pdb code 2ior). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total only one binding site of Magnesium was determined in the Crystal Structure of the N-Terminal Domain of Htpg, the Escherichia Coli HSP90, Bound to Adp, PDB code: 2ior:
In total only one binding site of Magnesium was determined in the Crystal Structure of the N-Terminal Domain of Htpg, the Escherichia Coli HSP90, Bound to Adp, PDB code: 2ior:
Magnesium binding site 1 out of 1 in 2ior
Go back to
Magnesium binding site 1 out
of 1 in the Crystal Structure of the N-Terminal Domain of Htpg, the Escherichia Coli HSP90, Bound to Adp
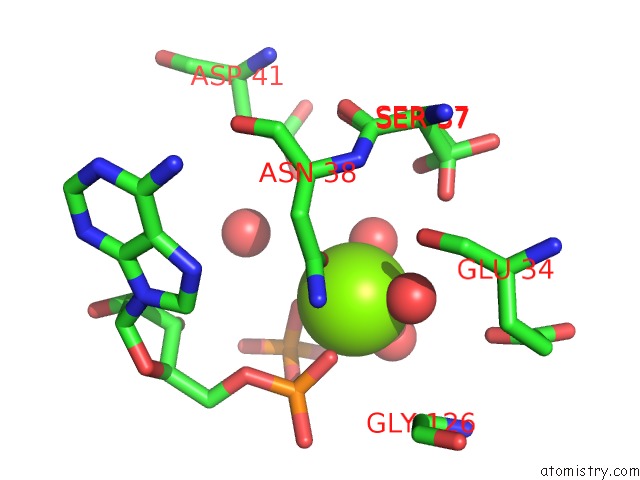
Mono view
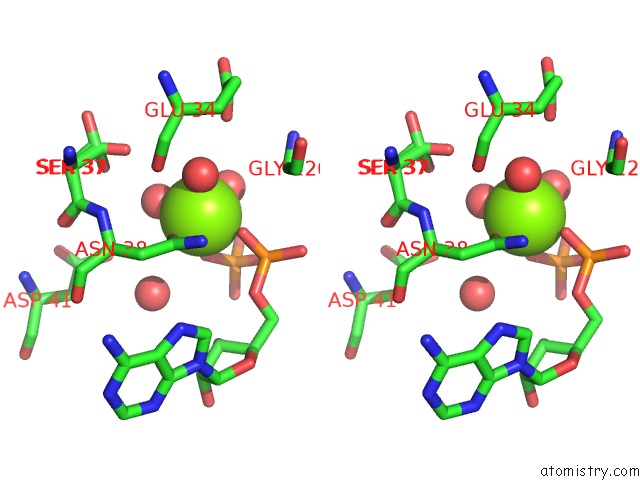
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Crystal Structure of the N-Terminal Domain of Htpg, the Escherichia Coli HSP90, Bound to Adp within 5.0Å range:
|
Reference:
A.K.Shiau,
S.F.Harris,
D.R.Southworth,
D.A.Agard.
Structural Analysis of E. Coli HSP90 Reveals Dramatic Nucleotide-Dependent Conformational Rearrangements. Cell(Cambridge,Mass.) V. 127 329 2006.
ISSN: ISSN 0092-8674
PubMed: 17055434
DOI: 10.1016/J.CELL.2006.09.027
Page generated: Sun Aug 10 11:38:55 2025
ISSN: ISSN 0092-8674
PubMed: 17055434
DOI: 10.1016/J.CELL.2006.09.027
Last articles
Mg in 2UUCMg in 2UX5
Mg in 2UX4
Mg in 2UX3
Mg in 2UWW
Mg in 2UWV
Mg in 2UWU
Mg in 2UWT
Mg in 2UUA
Mg in 2UWS