Magnesium »
PDB 2jga-2mse »
2m2u »
Magnesium in PDB 2m2u: Binary Complex of African Swine Fever Virus Pol X with Mgdgtp
Enzymatic activity of Binary Complex of African Swine Fever Virus Pol X with Mgdgtp
All present enzymatic activity of Binary Complex of African Swine Fever Virus Pol X with Mgdgtp:
2.7.7.7;
2.7.7.7;
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Binary Complex of African Swine Fever Virus Pol X with Mgdgtp
(pdb code 2m2u). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 2 binding sites of Magnesium where determined in the Binary Complex of African Swine Fever Virus Pol X with Mgdgtp, PDB code: 2m2u:
Jump to Magnesium binding site number: 1; 2;
In total 2 binding sites of Magnesium where determined in the Binary Complex of African Swine Fever Virus Pol X with Mgdgtp, PDB code: 2m2u:
Jump to Magnesium binding site number: 1; 2;
Magnesium binding site 1 out of 2 in 2m2u
Go back to
Magnesium binding site 1 out
of 2 in the Binary Complex of African Swine Fever Virus Pol X with Mgdgtp
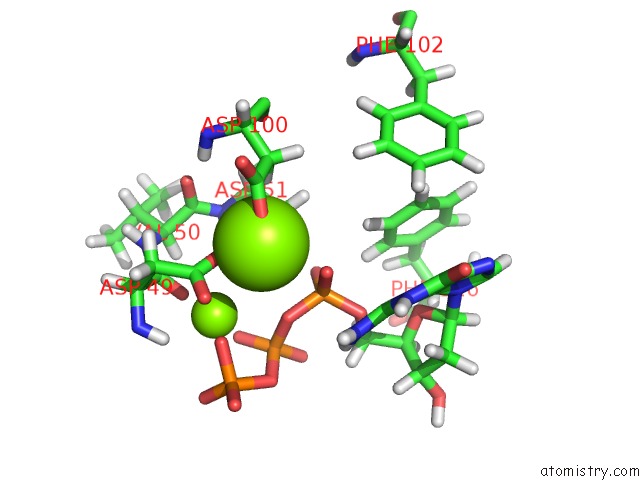
Mono view
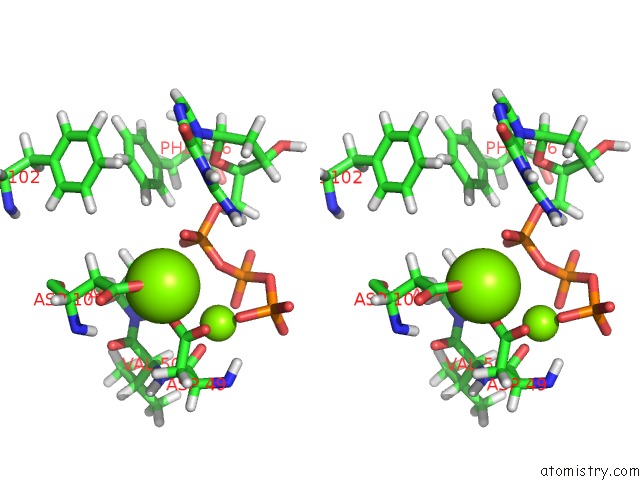
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Binary Complex of African Swine Fever Virus Pol X with Mgdgtp within 5.0Å range:
|
Magnesium binding site 2 out of 2 in 2m2u
Go back to
Magnesium binding site 2 out
of 2 in the Binary Complex of African Swine Fever Virus Pol X with Mgdgtp
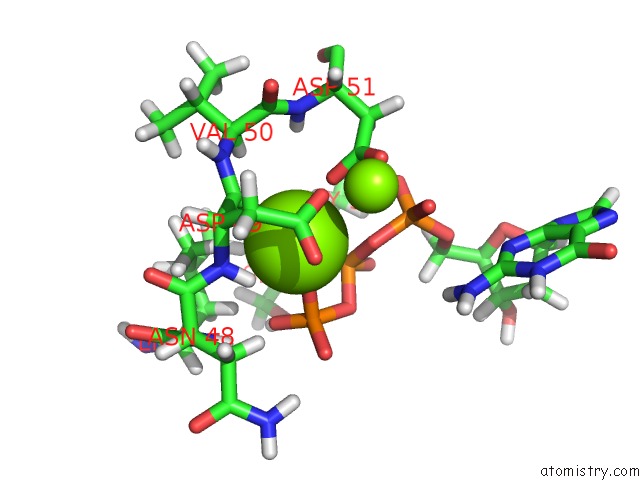
Mono view
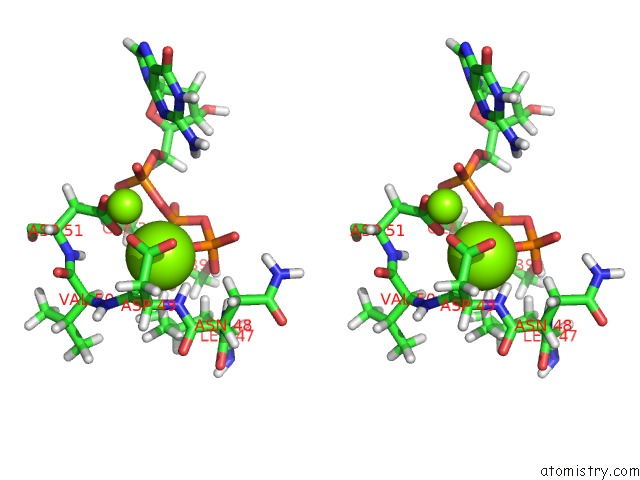
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Binary Complex of African Swine Fever Virus Pol X with Mgdgtp within 5.0Å range:
|
Reference:
W.J.Wu,
M.I.Su,
J.L.Wu,
S.Kumar,
L.H.Lim,
C.W.Wang,
F.H.Nelissen,
M.C.Chen,
J.F.Doreleijers,
S.S.Wijmenga,
M.D.Tsai.
How A Low-Fidelity Dna Polymerase Chooses Non-Watson-Crick From Watson-Crick Incorporation. J.Am.Chem.Soc. 2014.
ISSN: ESSN 1520-5126
PubMed: 24617852
DOI: 10.1021/JA4102375
Page generated: Wed Aug 14 00:51:43 2024
ISSN: ESSN 1520-5126
PubMed: 24617852
DOI: 10.1021/JA4102375
Last articles
Ca in 2Q16Ca in 2Q0R
Ca in 2PYH
Ca in 2Q0U
Ca in 2PY2
Ca in 2PZ0
Ca in 2PYZ
Ca in 2PYG
Ca in 2PWF
Ca in 2PWH