Magnesium »
PDB 2jga-2mse »
2msd »
Magnesium in PDB 2msd: uc(Nmr) Data-Driven Model of Gtpase Kras-Gnp Tethered to A Lipid-Bilayer Nanodisc
Magnesium Binding Sites:
The binding sites of Magnesium atom in the uc(Nmr) Data-Driven Model of Gtpase Kras-Gnp Tethered to A Lipid-Bilayer Nanodisc
(pdb code 2msd). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total only one binding site of Magnesium was determined in the uc(Nmr) Data-Driven Model of Gtpase Kras-Gnp Tethered to A Lipid-Bilayer Nanodisc, PDB code: 2msd:
In total only one binding site of Magnesium was determined in the uc(Nmr) Data-Driven Model of Gtpase Kras-Gnp Tethered to A Lipid-Bilayer Nanodisc, PDB code: 2msd:
Magnesium binding site 1 out of 1 in 2msd
Go back to
Magnesium binding site 1 out
of 1 in the uc(Nmr) Data-Driven Model of Gtpase Kras-Gnp Tethered to A Lipid-Bilayer Nanodisc

Mono view
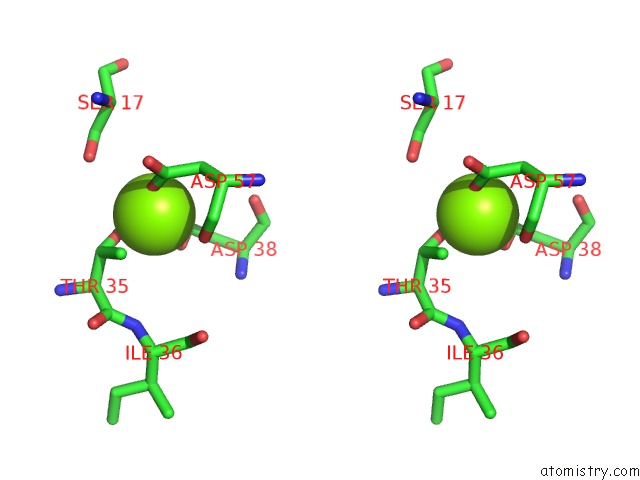
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of uc(Nmr) Data-Driven Model of Gtpase Kras-Gnp Tethered to A Lipid-Bilayer Nanodisc within 5.0Å range:
|
Reference:
M.T.Mazhab-Jafari,
C.B.Marshall,
M.J.Smith,
G.M.Gasmi-Seabrook,
P.B.Stathopulos,
F.Inagaki,
L.E.Kay,
B.G.Neel,
M.Ikura.
Oncogenic and Rasopathy-Associated K-Ras Mutations Relieve Membrane-Dependent Occlusion of the Effector-Binding Site. Proc.Natl.Acad.Sci.Usa V. 112 6625 2015.
ISSN: ISSN 0027-8424
PubMed: 25941399
DOI: 10.1073/PNAS.1419895112
Page generated: Wed Aug 14 00:52:20 2024
ISSN: ISSN 0027-8424
PubMed: 25941399
DOI: 10.1073/PNAS.1419895112
Last articles
Zn in 9MJ5Zn in 9HNW
Zn in 9G0L
Zn in 9FNE
Zn in 9DZN
Zn in 9E0I
Zn in 9D32
Zn in 9DAK
Zn in 8ZXC
Zn in 8ZUF