Magnesium »
PDB 2ppq-2q0e »
2ps8 »
Magnesium in PDB 2ps8: Y295F Trichodiene Synthase: Complex with Mg and Pyrophosphate
Enzymatic activity of Y295F Trichodiene Synthase: Complex with Mg and Pyrophosphate
All present enzymatic activity of Y295F Trichodiene Synthase: Complex with Mg and Pyrophosphate:
4.2.3.6;
4.2.3.6;
Protein crystallography data
The structure of Y295F Trichodiene Synthase: Complex with Mg and Pyrophosphate, PDB code: 2ps8
was solved by
L.S.Vedula,
D.E.Cane,
D.W.Christianson,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 50.00 / 2.67 |
| Space group | P 31 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 122.272, 122.272, 150.474, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 20.3 / 24.5 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Y295F Trichodiene Synthase: Complex with Mg and Pyrophosphate
(pdb code 2ps8). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 3 binding sites of Magnesium where determined in the Y295F Trichodiene Synthase: Complex with Mg and Pyrophosphate, PDB code: 2ps8:
Jump to Magnesium binding site number: 1; 2; 3;
In total 3 binding sites of Magnesium where determined in the Y295F Trichodiene Synthase: Complex with Mg and Pyrophosphate, PDB code: 2ps8:
Jump to Magnesium binding site number: 1; 2; 3;
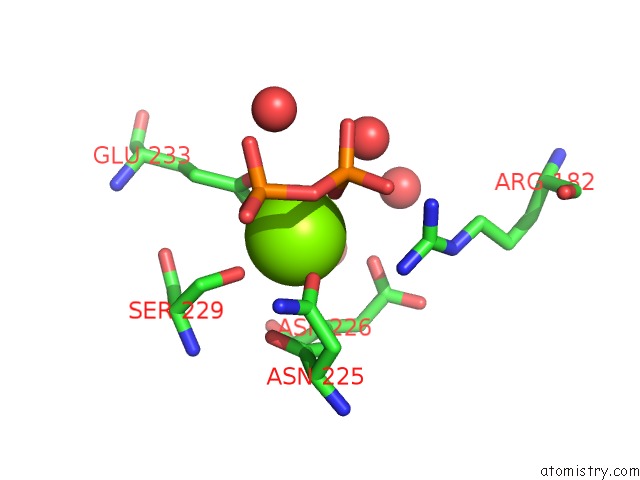
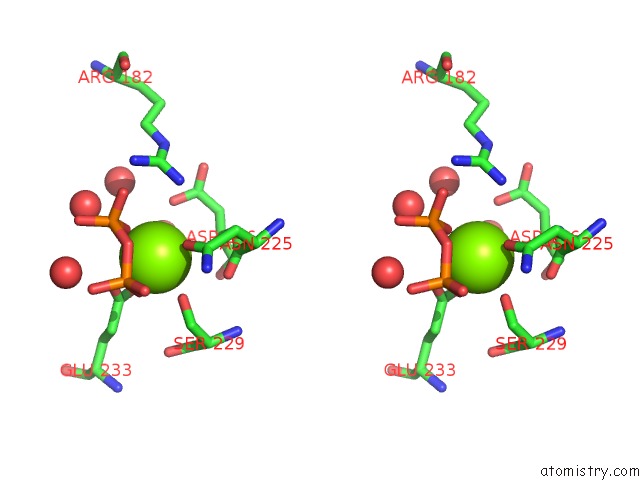
Magnesium binding site 1 out of 3 in 2ps8
Go back to
Magnesium binding site 1 out
of 3 in the Y295F Trichodiene Synthase: Complex with Mg and Pyrophosphate
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Y295F Trichodiene Synthase: Complex with Mg and Pyrophosphate within 5.0Å range:
|
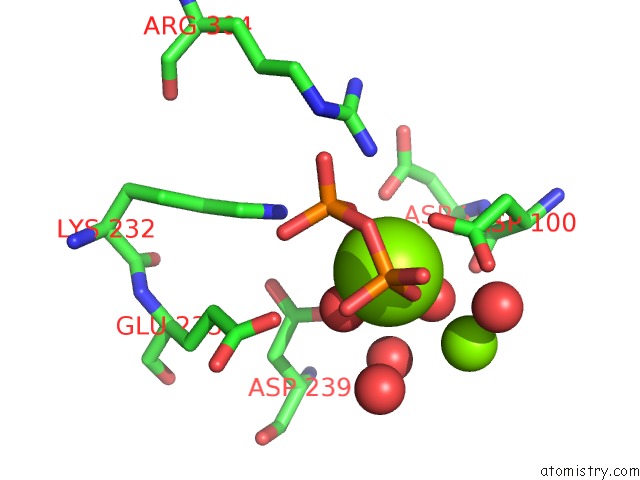
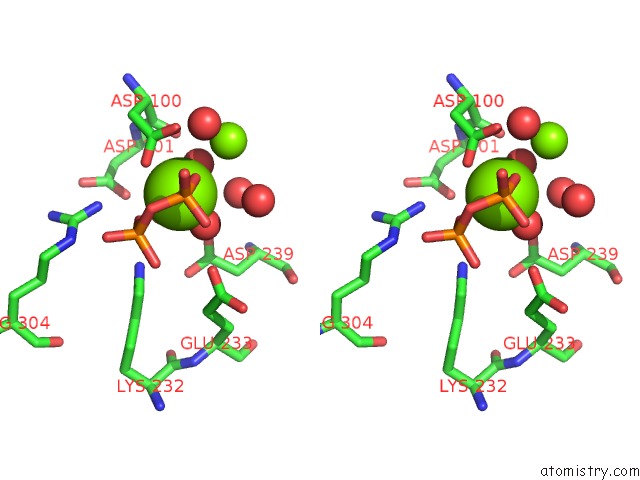
Magnesium binding site 2 out of 3 in 2ps8
Go back to
Magnesium binding site 2 out
of 3 in the Y295F Trichodiene Synthase: Complex with Mg and Pyrophosphate
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Y295F Trichodiene Synthase: Complex with Mg and Pyrophosphate within 5.0Å range:
|
Magnesium binding site 3 out of 3 in 2ps8
Go back to
Magnesium binding site 3 out
of 3 in the Y295F Trichodiene Synthase: Complex with Mg and Pyrophosphate
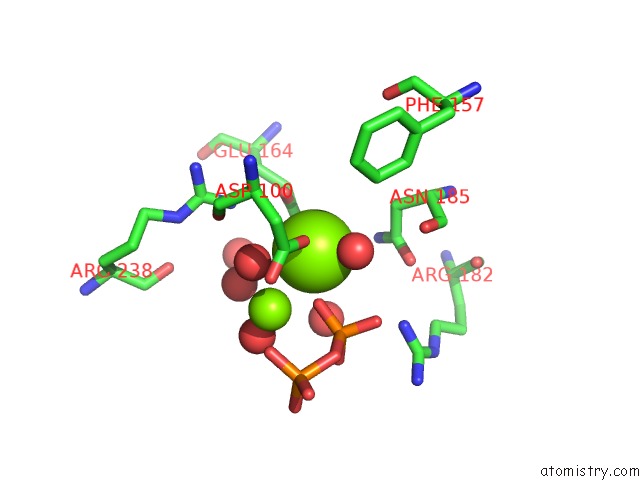
Mono view
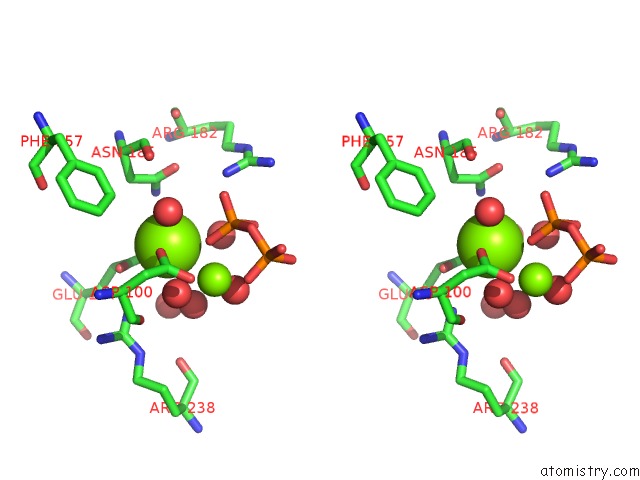
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 3 of Y295F Trichodiene Synthase: Complex with Mg and Pyrophosphate within 5.0Å range:
|
Reference:
L.S.Vedula,
J.Jiang,
T.Zakharian,
D.E.Cane,
D.W.Christianson.
Structural and Mechanistic Analysis of Trichodiene Synthase Using Site-Directed Mutagenesis: Probing the Catalytic Function of Tyrosine-295 and the Asparagine-225/Serine-229/Glutamate-233-MG2+B Motif. Arch.Biochem.Biophys. V. 469 184 2008.
ISSN: ISSN 0003-9861
PubMed: 17996718
DOI: 10.1016/J.ABB.2007.10.015
Page generated: Wed Aug 14 02:16:57 2024
ISSN: ISSN 0003-9861
PubMed: 17996718
DOI: 10.1016/J.ABB.2007.10.015
Last articles
Cl in 5XQECl in 5XQD
Cl in 5XPE
Cl in 5XNL
Cl in 5XP7
Cl in 5XNQ
Cl in 5XNV
Cl in 5XNA
Cl in 5XMV
Cl in 5XII