Magnesium »
PDB 2q9y-2qrf »
2qoj »
Magnesium in PDB 2qoj: Coevolution of A Homing Endonuclease and Its Host Target Sequence
Protein crystallography data
The structure of Coevolution of A Homing Endonuclease and Its Host Target Sequence, PDB code: 2qoj
was solved by
M.Scalley-Kim,
A.Mcconnell Smith,
B.L.Stoddard,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 29.30 / 2.40 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 60.353, 72.701, 61.100, 90.00, 103.88, 90.00 |
| R / Rfree (%) | 21.6 / 27.6 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Coevolution of A Homing Endonuclease and Its Host Target Sequence
(pdb code 2qoj). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 3 binding sites of Magnesium where determined in the Coevolution of A Homing Endonuclease and Its Host Target Sequence, PDB code: 2qoj:
Jump to Magnesium binding site number: 1; 2; 3;
In total 3 binding sites of Magnesium where determined in the Coevolution of A Homing Endonuclease and Its Host Target Sequence, PDB code: 2qoj:
Jump to Magnesium binding site number: 1; 2; 3;
Magnesium binding site 1 out of 3 in 2qoj
Go back to
Magnesium binding site 1 out
of 3 in the Coevolution of A Homing Endonuclease and Its Host Target Sequence

Mono view
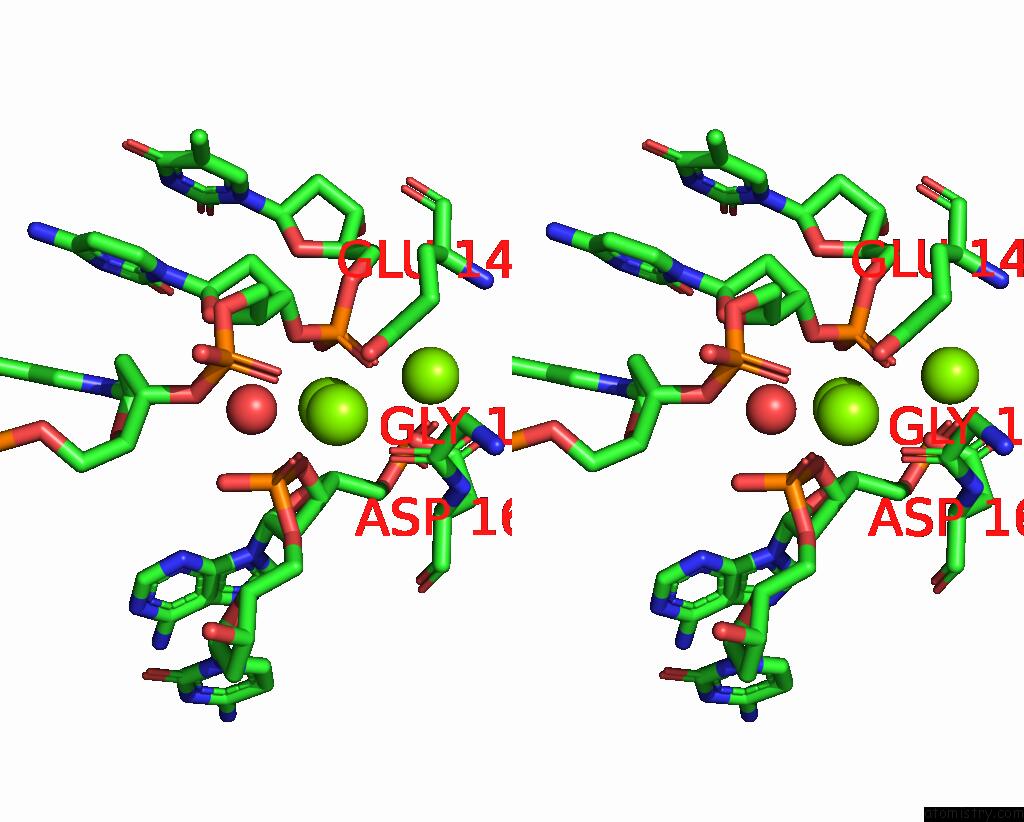
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Coevolution of A Homing Endonuclease and Its Host Target Sequence within 5.0Å range:
|
Magnesium binding site 2 out of 3 in 2qoj
Go back to
Magnesium binding site 2 out
of 3 in the Coevolution of A Homing Endonuclease and Its Host Target Sequence

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Coevolution of A Homing Endonuclease and Its Host Target Sequence within 5.0Å range:
|
Magnesium binding site 3 out of 3 in 2qoj
Go back to
Magnesium binding site 3 out
of 3 in the Coevolution of A Homing Endonuclease and Its Host Target Sequence
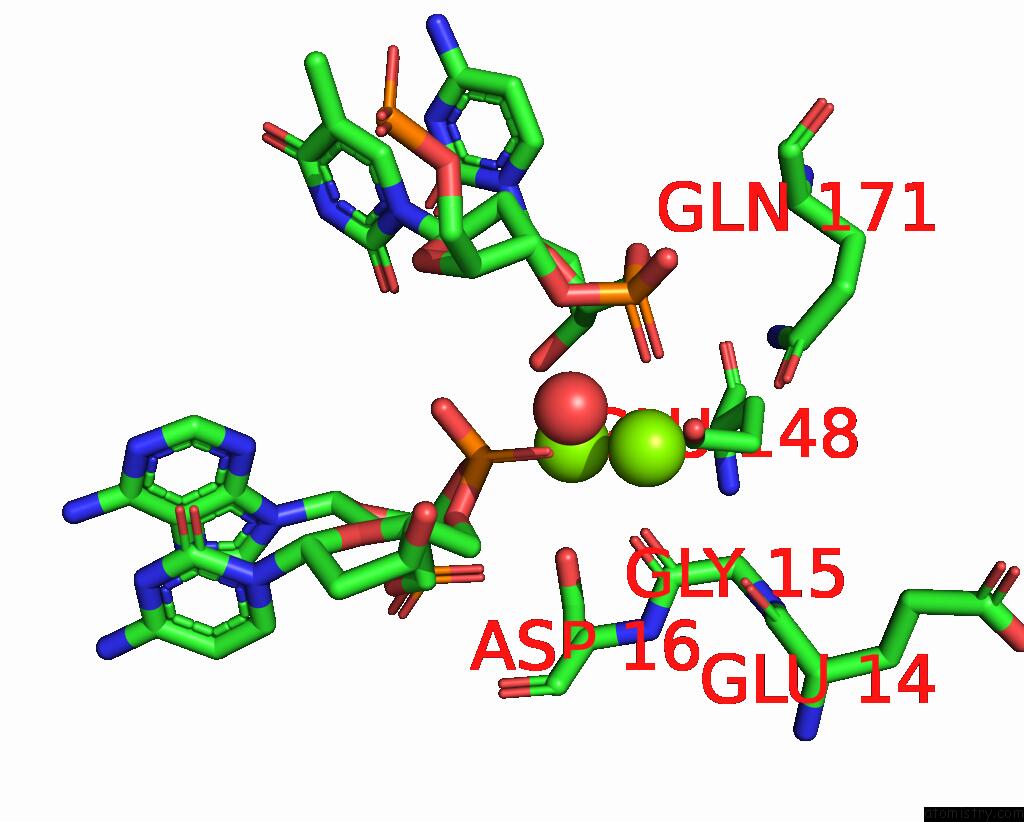
Mono view
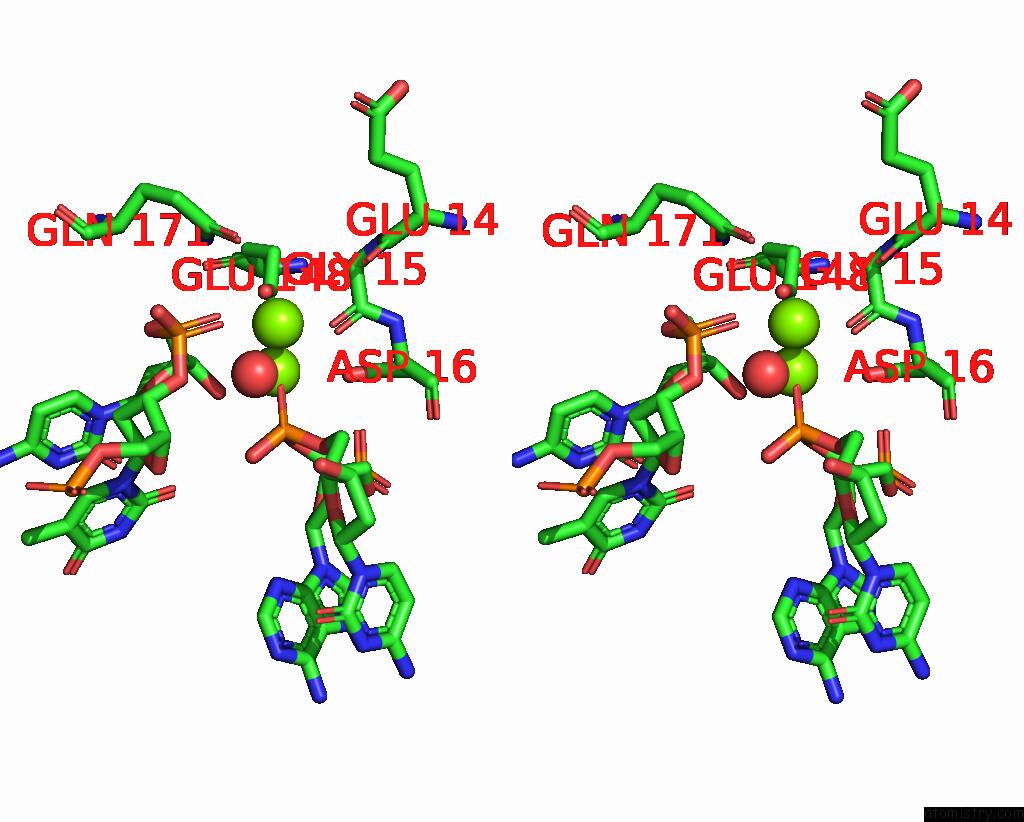
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 3 of Coevolution of A Homing Endonuclease and Its Host Target Sequence within 5.0Å range:
|
Reference:
M.Scalley-Kim,
A.Mcconnell-Smith,
B.L.Stoddard.
Coevolution of A Homing Endonuclease and Its Host Target Sequence. J.Mol.Biol. V. 372 1305 2007.
ISSN: ISSN 0022-2836
PubMed: 17720189
DOI: 10.1016/J.JMB.2007.07.052
Page generated: Sun Aug 10 13:25:04 2025
ISSN: ISSN 0022-2836
PubMed: 17720189
DOI: 10.1016/J.JMB.2007.07.052
Last articles
Mg in 3CB3Mg in 3CC6
Mg in 3C9U
Mg in 3CBT
Mg in 3CBQ
Mg in 3CBG
Mg in 3CBE
Mg in 3C9T
Mg in 3CB9
Mg in 3CAW