Magnesium »
PDB 2xzv-2yaz »
2y4g »
Magnesium in PDB 2y4g: Structure of the Tirandamycin-Bound Fad-Dependent Tirandamycin Oxidase Taml in P212121 Space Group
Protein crystallography data
The structure of Structure of the Tirandamycin-Bound Fad-Dependent Tirandamycin Oxidase Taml in P212121 Space Group, PDB code: 2y4g
was solved by
J.C.Carlson,
S.Li,
S.S.Gunatilleke,
Y.Anzai,
D.A.Burr,
L.M.Podust,
D.H.Sherman,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 134.54 / 2.03 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 64.020, 130.400, 134.540, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 15.886 / 23.566 |
Other elements in 2y4g:
The structure of Structure of the Tirandamycin-Bound Fad-Dependent Tirandamycin Oxidase Taml in P212121 Space Group also contains other interesting chemical elements:
| Chlorine | (Cl) | 5 atoms |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Structure of the Tirandamycin-Bound Fad-Dependent Tirandamycin Oxidase Taml in P212121 Space Group
(pdb code 2y4g). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 4 binding sites of Magnesium where determined in the Structure of the Tirandamycin-Bound Fad-Dependent Tirandamycin Oxidase Taml in P212121 Space Group, PDB code: 2y4g:
Jump to Magnesium binding site number: 1; 2; 3; 4;
In total 4 binding sites of Magnesium where determined in the Structure of the Tirandamycin-Bound Fad-Dependent Tirandamycin Oxidase Taml in P212121 Space Group, PDB code: 2y4g:
Jump to Magnesium binding site number: 1; 2; 3; 4;
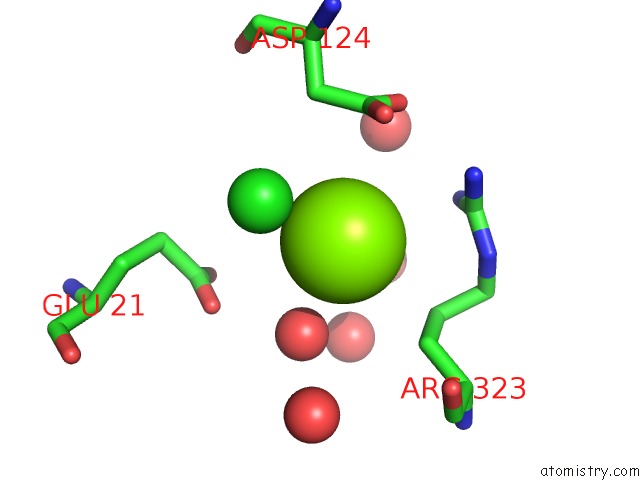
Magnesium binding site 1 out of 4 in 2y4g
Go back to
Magnesium binding site 1 out
of 4 in the Structure of the Tirandamycin-Bound Fad-Dependent Tirandamycin Oxidase Taml in P212121 Space Group
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Structure of the Tirandamycin-Bound Fad-Dependent Tirandamycin Oxidase Taml in P212121 Space Group within 5.0Å range:
|
Magnesium binding site 2 out of 4 in 2y4g
Go back to
Magnesium binding site 2 out
of 4 in the Structure of the Tirandamycin-Bound Fad-Dependent Tirandamycin Oxidase Taml in P212121 Space Group

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Structure of the Tirandamycin-Bound Fad-Dependent Tirandamycin Oxidase Taml in P212121 Space Group within 5.0Å range:
|
Magnesium binding site 3 out of 4 in 2y4g
Go back to
Magnesium binding site 3 out
of 4 in the Structure of the Tirandamycin-Bound Fad-Dependent Tirandamycin Oxidase Taml in P212121 Space Group

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 3 of Structure of the Tirandamycin-Bound Fad-Dependent Tirandamycin Oxidase Taml in P212121 Space Group within 5.0Å range:
|
Magnesium binding site 4 out of 4 in 2y4g
Go back to
Magnesium binding site 4 out
of 4 in the Structure of the Tirandamycin-Bound Fad-Dependent Tirandamycin Oxidase Taml in P212121 Space Group

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 4 of Structure of the Tirandamycin-Bound Fad-Dependent Tirandamycin Oxidase Taml in P212121 Space Group within 5.0Å range:
|
Reference:
J.C.Carlson,
S.Li,
S.S.Gunatilleke,
Y.Anzai,
D.A.Burr,
L.M.Podust,
D.H.Sherman.
Tirandamycin Biosynthesis Is Mediated By Co-Dependent Oxidative Enzymes Nat.Chem V. 3 628 2011.
ISSN: ISSN 1755-4330
PubMed: 21778983
DOI: 10.1038/NCHEM.1087
Page generated: Sun Aug 10 16:37:21 2025
ISSN: ISSN 1755-4330
PubMed: 21778983
DOI: 10.1038/NCHEM.1087
Last articles
Mg in 3CRRMg in 3CRQ
Mg in 3CRC
Mg in 3CR3
Mg in 3CQ3
Mg in 3CQD
Mg in 3CR1
Mg in 3CME
Mg in 3CMA
Mg in 3CP6