Magnesium »
PDB 2ynj-2z4w »
2ynm »
Magnesium in PDB 2ynm: Structure of the ADPXALF3-Stabilized Transition State of the Nitrogenase-Like Dark-Operative Protochlorophyllide Oxidoreductase Complex From Prochlorococcus Marinus with Its Substrate Protochlorophyllide A
Enzymatic activity of Structure of the ADPXALF3-Stabilized Transition State of the Nitrogenase-Like Dark-Operative Protochlorophyllide Oxidoreductase Complex From Prochlorococcus Marinus with Its Substrate Protochlorophyllide A
All present enzymatic activity of Structure of the ADPXALF3-Stabilized Transition State of the Nitrogenase-Like Dark-Operative Protochlorophyllide Oxidoreductase Complex From Prochlorococcus Marinus with Its Substrate Protochlorophyllide A:
1.3.7.7;
1.3.7.7;
Protein crystallography data
The structure of Structure of the ADPXALF3-Stabilized Transition State of the Nitrogenase-Like Dark-Operative Protochlorophyllide Oxidoreductase Complex From Prochlorococcus Marinus with Its Substrate Protochlorophyllide A, PDB code: 2ynm
was solved by
J.Krausze,
C.Lange,
D.W.Heinz,
J.Moser,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 34.510 / 2.10 |
| Space group | C 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 308.400, 74.110, 74.230, 90.00, 91.24, 90.00 |
| R / Rfree (%) | 19.89 / 23.31 |
Other elements in 2ynm:
The structure of Structure of the ADPXALF3-Stabilized Transition State of the Nitrogenase-Like Dark-Operative Protochlorophyllide Oxidoreductase Complex From Prochlorococcus Marinus with Its Substrate Protochlorophyllide A also contains other interesting chemical elements:
| Fluorine | (F) | 6 atoms |
| Aluminium | (Al) | 2 atoms |
| Potassium | (K) | 1 atom |
| Iron | (Fe) | 8 atoms |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Structure of the ADPXALF3-Stabilized Transition State of the Nitrogenase-Like Dark-Operative Protochlorophyllide Oxidoreductase Complex From Prochlorococcus Marinus with Its Substrate Protochlorophyllide A
(pdb code 2ynm). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 3 binding sites of Magnesium where determined in the Structure of the ADPXALF3-Stabilized Transition State of the Nitrogenase-Like Dark-Operative Protochlorophyllide Oxidoreductase Complex From Prochlorococcus Marinus with Its Substrate Protochlorophyllide A, PDB code: 2ynm:
Jump to Magnesium binding site number: 1; 2; 3;
In total 3 binding sites of Magnesium where determined in the Structure of the ADPXALF3-Stabilized Transition State of the Nitrogenase-Like Dark-Operative Protochlorophyllide Oxidoreductase Complex From Prochlorococcus Marinus with Its Substrate Protochlorophyllide A, PDB code: 2ynm:
Jump to Magnesium binding site number: 1; 2; 3;
Magnesium binding site 1 out of 3 in 2ynm
Go back to
Magnesium binding site 1 out
of 3 in the Structure of the ADPXALF3-Stabilized Transition State of the Nitrogenase-Like Dark-Operative Protochlorophyllide Oxidoreductase Complex From Prochlorococcus Marinus with Its Substrate Protochlorophyllide A
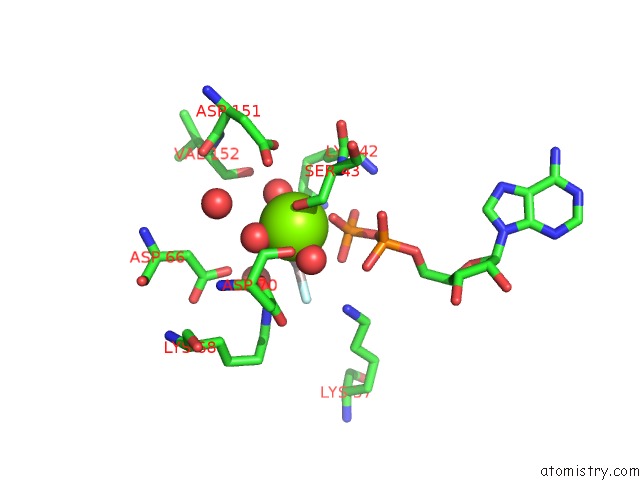
Mono view
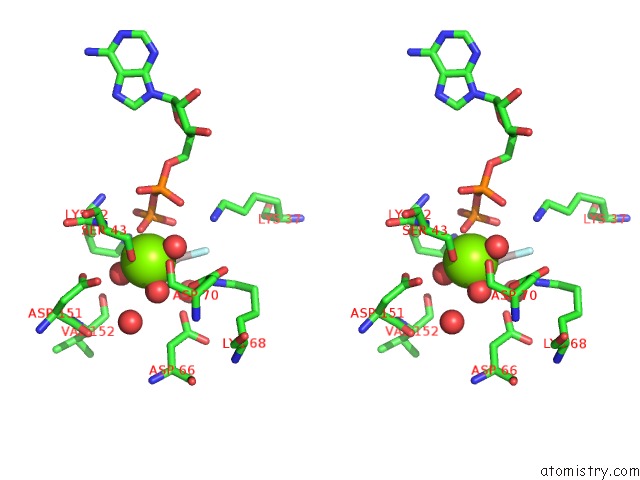
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Structure of the ADPXALF3-Stabilized Transition State of the Nitrogenase-Like Dark-Operative Protochlorophyllide Oxidoreductase Complex From Prochlorococcus Marinus with Its Substrate Protochlorophyllide A within 5.0Å range:
|
Magnesium binding site 2 out of 3 in 2ynm
Go back to
Magnesium binding site 2 out
of 3 in the Structure of the ADPXALF3-Stabilized Transition State of the Nitrogenase-Like Dark-Operative Protochlorophyllide Oxidoreductase Complex From Prochlorococcus Marinus with Its Substrate Protochlorophyllide A
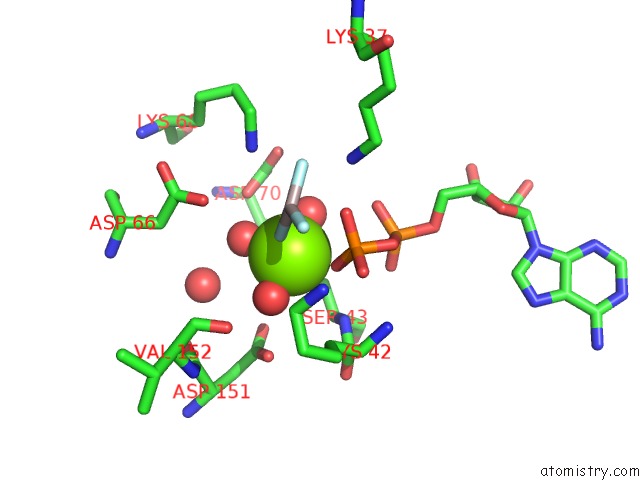
Mono view
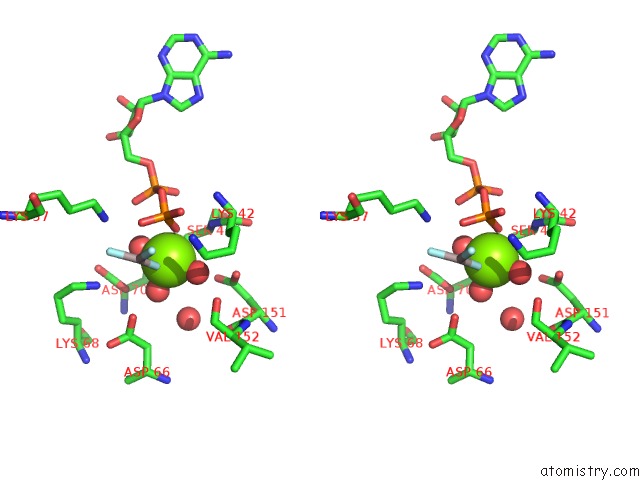
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Structure of the ADPXALF3-Stabilized Transition State of the Nitrogenase-Like Dark-Operative Protochlorophyllide Oxidoreductase Complex From Prochlorococcus Marinus with Its Substrate Protochlorophyllide A within 5.0Å range:
|
Magnesium binding site 3 out of 3 in 2ynm
Go back to
Magnesium binding site 3 out
of 3 in the Structure of the ADPXALF3-Stabilized Transition State of the Nitrogenase-Like Dark-Operative Protochlorophyllide Oxidoreductase Complex From Prochlorococcus Marinus with Its Substrate Protochlorophyllide A
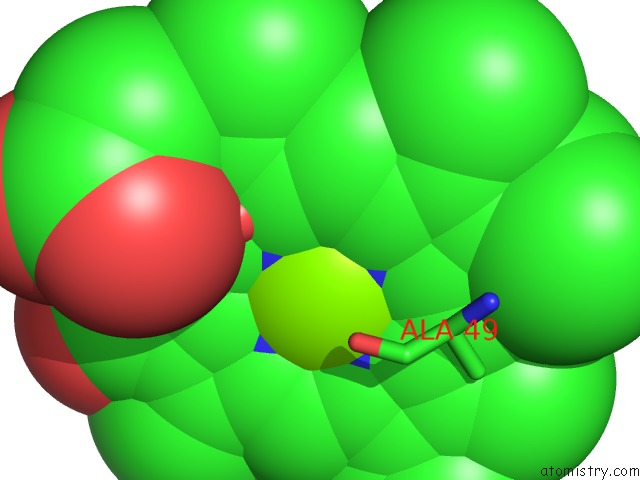
Mono view
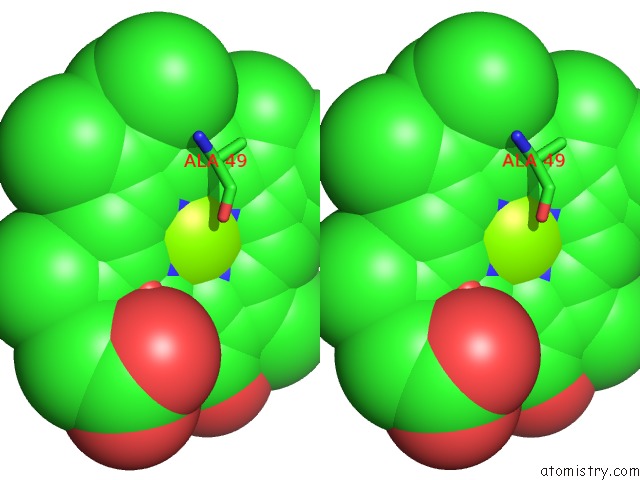
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 3 of Structure of the ADPXALF3-Stabilized Transition State of the Nitrogenase-Like Dark-Operative Protochlorophyllide Oxidoreductase Complex From Prochlorococcus Marinus with Its Substrate Protochlorophyllide A within 5.0Å range:
|
Reference:
J.Moser,
C.Lange,
J.Krausze,
J.Rebelein,
W.Schubert,
M.W.Ribbe,
D.W.Heinz,
D.Jahn.
Structure of Adp-Aluminium Fluoride-Stabilized Protochlorophyllide Oxidoreductase Complex. Proc.Natl.Acad.Sci.Usa V. 110 2094 2013.
ISSN: ISSN 0027-8424
PubMed: 23341615
DOI: 10.1073/PNAS.1218303110
Page generated: Wed Aug 14 07:38:21 2024
ISSN: ISSN 0027-8424
PubMed: 23341615
DOI: 10.1073/PNAS.1218303110
Last articles
Cl in 7XI2Cl in 7XHQ
Cl in 7XI0
Cl in 7XHR
Cl in 7XGN
Cl in 7XHH
Cl in 7XGM
Cl in 7XHK
Cl in 7XGL
Cl in 7XGI