Magnesium »
PDB 3bdh-3bre »
3bjy »
Magnesium in PDB 3bjy: Catalytic Core of REV1 in Complex with Dna (Modified Template Guanine) and Incoming Nucleotide
Protein crystallography data
The structure of Catalytic Core of REV1 in Complex with Dna (Modified Template Guanine) and Incoming Nucleotide, PDB code: 3bjy
was solved by
D.T.Nair,
R.E.Johnson,
L.Prakash,
S.Prakash,
A.K.Aggarwal,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 50.00 / 2.41 |
| Space group | C 2 2 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 181.062, 199.578, 55.677, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 23.1 / 24.1 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Catalytic Core of REV1 in Complex with Dna (Modified Template Guanine) and Incoming Nucleotide
(pdb code 3bjy). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 2 binding sites of Magnesium where determined in the Catalytic Core of REV1 in Complex with Dna (Modified Template Guanine) and Incoming Nucleotide, PDB code: 3bjy:
Jump to Magnesium binding site number: 1; 2;
In total 2 binding sites of Magnesium where determined in the Catalytic Core of REV1 in Complex with Dna (Modified Template Guanine) and Incoming Nucleotide, PDB code: 3bjy:
Jump to Magnesium binding site number: 1; 2;
Magnesium binding site 1 out of 2 in 3bjy
Go back to
Magnesium binding site 1 out
of 2 in the Catalytic Core of REV1 in Complex with Dna (Modified Template Guanine) and Incoming Nucleotide
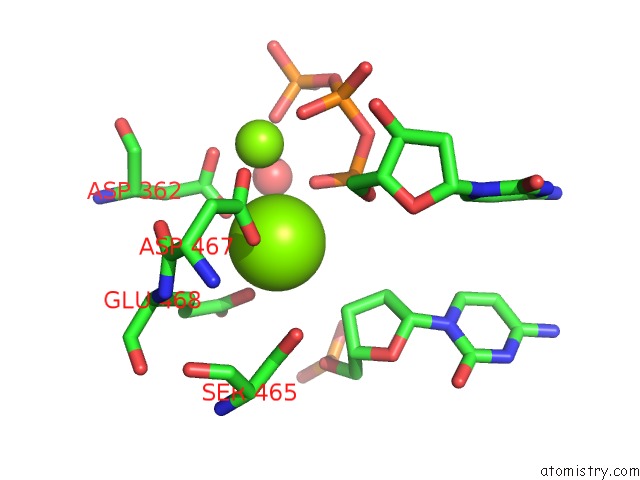
Mono view
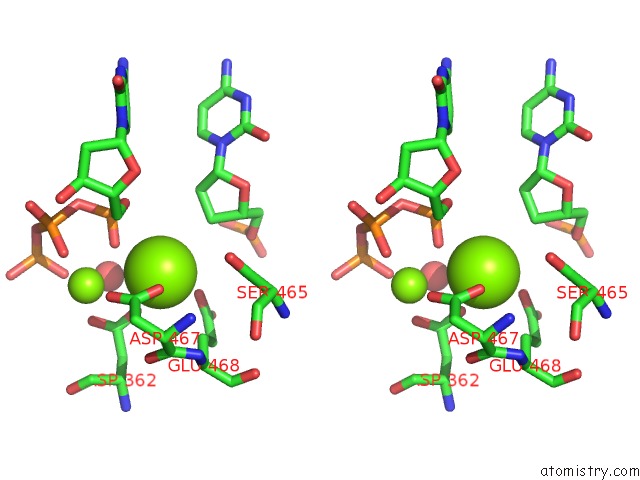
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Catalytic Core of REV1 in Complex with Dna (Modified Template Guanine) and Incoming Nucleotide within 5.0Å range:
|
Magnesium binding site 2 out of 2 in 3bjy
Go back to
Magnesium binding site 2 out
of 2 in the Catalytic Core of REV1 in Complex with Dna (Modified Template Guanine) and Incoming Nucleotide
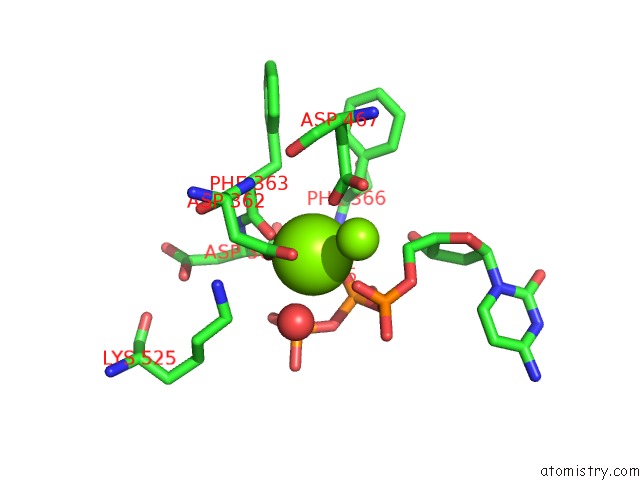
Mono view
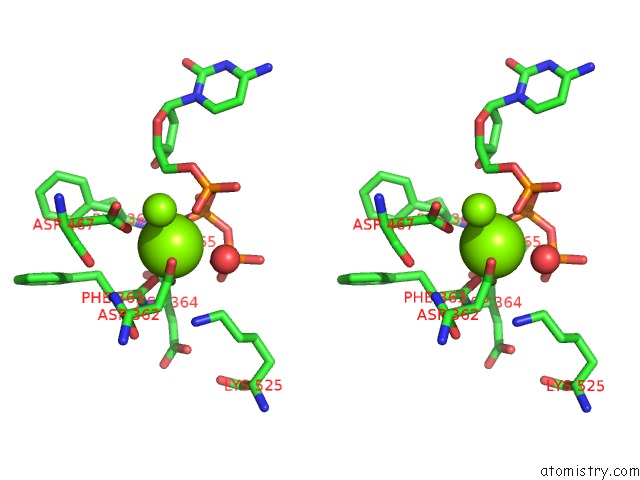
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Catalytic Core of REV1 in Complex with Dna (Modified Template Guanine) and Incoming Nucleotide within 5.0Å range:
|
Reference:
D.T.Nair,
R.E.Johnson,
L.Prakash,
S.Prakash,
A.K.Aggarwal.
Protein-Template-Directed Synthesis Across An Acrolein-Derived Dna Adduct By Yeast REV1 Dna Polymerase Structure V. 16 239 2008.
ISSN: ISSN 0969-2126
PubMed: 18275815
DOI: 10.1016/J.STR.2007.12.009
Page generated: Sun Aug 10 17:50:01 2025
ISSN: ISSN 0969-2126
PubMed: 18275815
DOI: 10.1016/J.STR.2007.12.009
Last articles
Mg in 6CGDMg in 6CFV
Mg in 6CFU
Mg in 6CFT
Mg in 6CEY
Mg in 6CFS
Mg in 6CFR
Mg in 6CFO
Mg in 6CER
Mg in 6CC3