Magnesium »
PDB 3dfy-3dts »
3dnt »
Magnesium in PDB 3dnt: Structures of Mdt Proteins
Protein crystallography data
The structure of Structures of Mdt Proteins, PDB code: 3dnt
was solved by
M.A.Schumacher,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 42.58 / 1.66 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 68.540, 84.090, 69.250, 90.00, 91.56, 90.00 |
| R / Rfree (%) | 18.4 / 21.7 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Structures of Mdt Proteins
(pdb code 3dnt). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 4 binding sites of Magnesium where determined in the Structures of Mdt Proteins, PDB code: 3dnt:
Jump to Magnesium binding site number: 1; 2; 3; 4;
In total 4 binding sites of Magnesium where determined in the Structures of Mdt Proteins, PDB code: 3dnt:
Jump to Magnesium binding site number: 1; 2; 3; 4;
Magnesium binding site 1 out of 4 in 3dnt
Go back to
Magnesium binding site 1 out
of 4 in the Structures of Mdt Proteins

Mono view
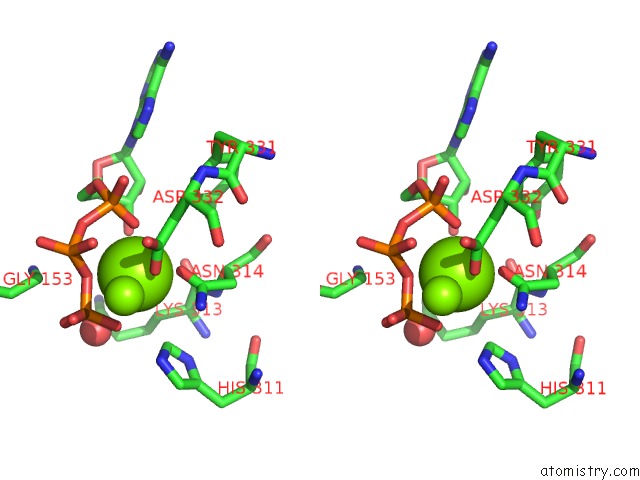
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Structures of Mdt Proteins within 5.0Å range:
|
Magnesium binding site 2 out of 4 in 3dnt
Go back to
Magnesium binding site 2 out
of 4 in the Structures of Mdt Proteins

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Structures of Mdt Proteins within 5.0Å range:
|
Magnesium binding site 3 out of 4 in 3dnt
Go back to
Magnesium binding site 3 out
of 4 in the Structures of Mdt Proteins

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 3 of Structures of Mdt Proteins within 5.0Å range:
|
Magnesium binding site 4 out of 4 in 3dnt
Go back to
Magnesium binding site 4 out
of 4 in the Structures of Mdt Proteins

Mono view
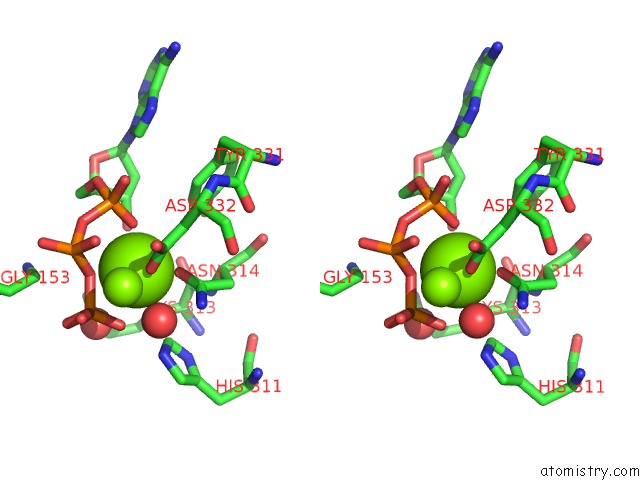
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 4 of Structures of Mdt Proteins within 5.0Å range:
|
Reference:
M.A.Schumacher,
K.M.Piro,
W.Xu,
S.Hansen,
K.Lewis,
R.G.Brennan.
Molecular Mechanisms of Hipa-Mediated Multidrug Tolerance and Its Neutralization By Hipb. Science V. 323 396 2009.
ISSN: ISSN 0036-8075
PubMed: 19150849
DOI: 10.1126/SCIENCE.1163806
Page generated: Wed Aug 14 12:35:08 2024
ISSN: ISSN 0036-8075
PubMed: 19150849
DOI: 10.1126/SCIENCE.1163806
Last articles
F in 4IVMF in 4IV2
F in 4IUI
F in 4IN4
F in 4IU7
F in 4ITI
F in 4IUE
F in 4IRU
F in 4ITJ
F in 4ISF