Magnesium »
PDB 3ism-3jaw »
3jap »
Magnesium in PDB 3jap: Structure of A Partial Yeast 48S Preinitiation Complex in Closed Conformation
Other elements in 3jap:
The structure of Structure of A Partial Yeast 48S Preinitiation Complex in Closed Conformation also contains other interesting chemical elements:
| Zinc | (Zn) | 4 atoms |
Magnesium Binding Sites:
Pages:
>>> Page 1 <<< Page 2, Binding sites: 11 - 20; Page 3, Binding sites: 21 - 30; Page 4, Binding sites: 31 - 40; Page 5, Binding sites: 41 - 50; Page 6, Binding sites: 51 - 60; Page 7, Binding sites: 61 - 70; Page 8, Binding sites: 71 - 80; Page 9, Binding sites: 81 - 81;Binding sites:
The binding sites of Magnesium atom in the Structure of A Partial Yeast 48S Preinitiation Complex in Closed Conformation (pdb code 3jap). This binding sites where shown within 5.0 Angstroms radius around Magnesium atom.In total 81 binding sites of Magnesium where determined in the Structure of A Partial Yeast 48S Preinitiation Complex in Closed Conformation, PDB code: 3jap:
Jump to Magnesium binding site number: 1; 2; 3; 4; 5; 6; 7; 8; 9; 10;
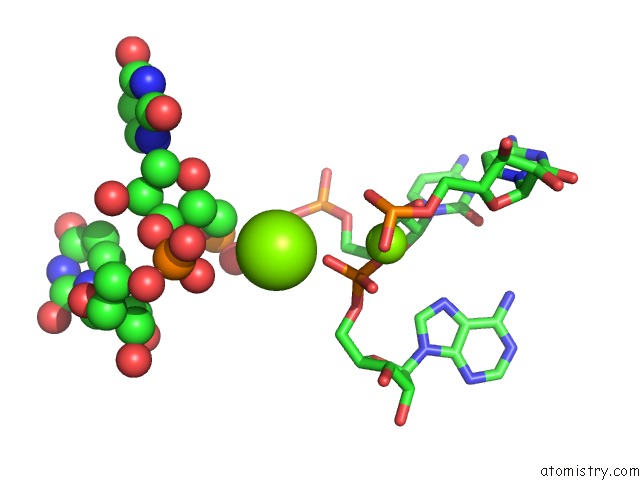
Magnesium binding site 1 out of 81 in 3jap
Go back to
Magnesium binding site 1 out
of 81 in the Structure of A Partial Yeast 48S Preinitiation Complex in Closed Conformation
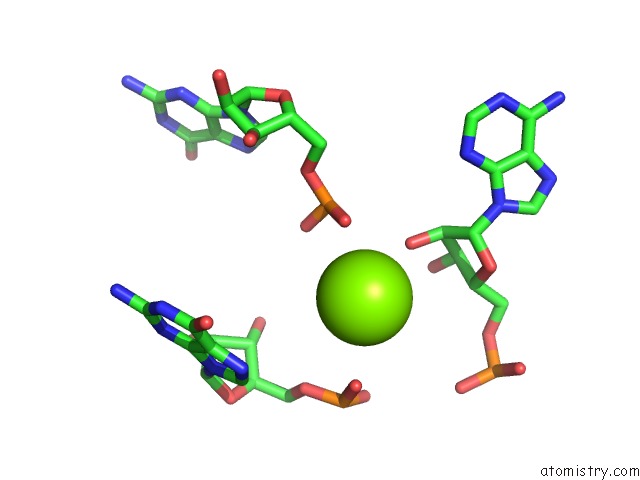
Mono view
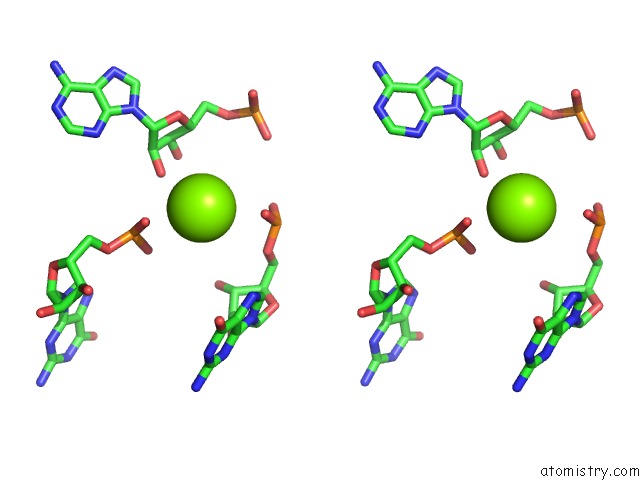
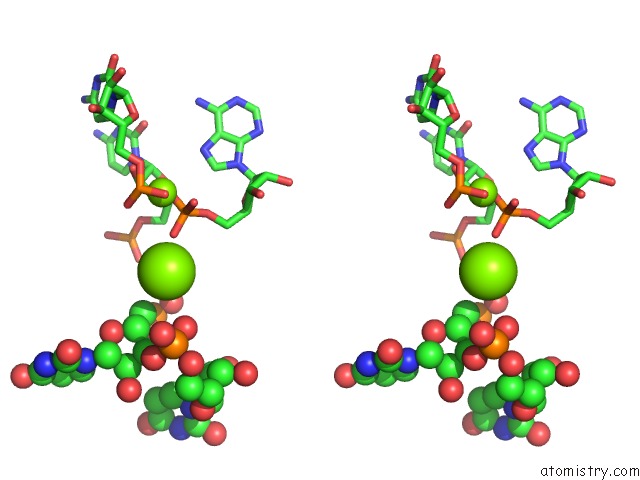
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Structure of A Partial Yeast 48S Preinitiation Complex in Closed Conformation within 5.0Å range:
|
Magnesium binding site 2 out of 81 in 3jap
Go back to
Magnesium binding site 2 out
of 81 in the Structure of A Partial Yeast 48S Preinitiation Complex in Closed Conformation
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Structure of A Partial Yeast 48S Preinitiation Complex in Closed Conformation within 5.0Å range:
|
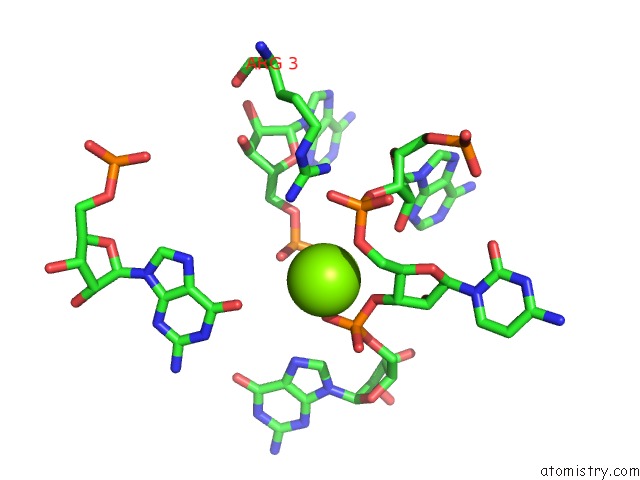
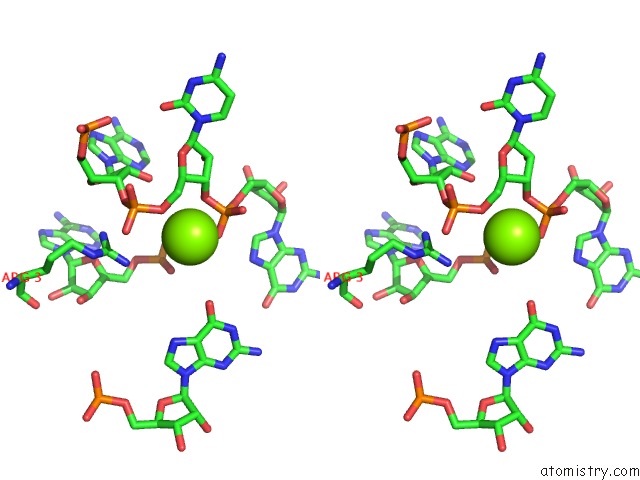
Magnesium binding site 3 out of 81 in 3jap
Go back to
Magnesium binding site 3 out
of 81 in the Structure of A Partial Yeast 48S Preinitiation Complex in Closed Conformation
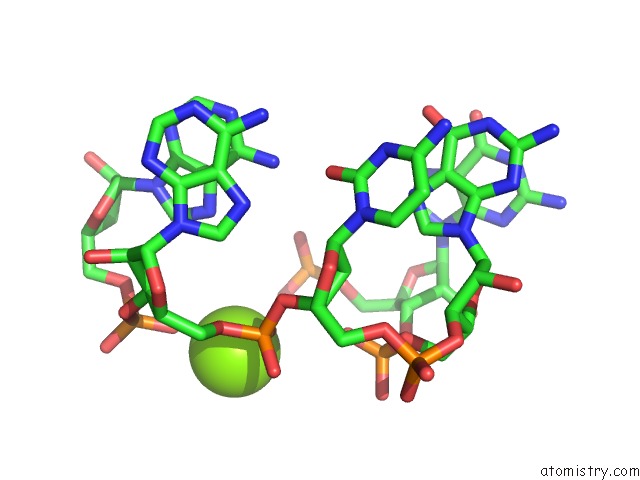
Mono view
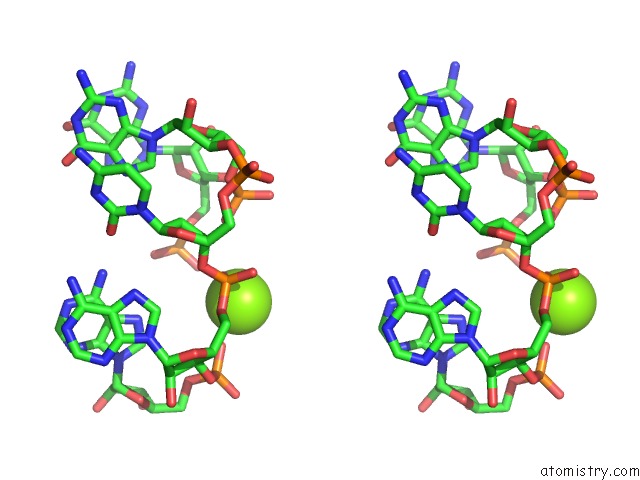
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 3 of Structure of A Partial Yeast 48S Preinitiation Complex in Closed Conformation within 5.0Å range:
|
Magnesium binding site 4 out of 81 in 3jap
Go back to
Magnesium binding site 4 out
of 81 in the Structure of A Partial Yeast 48S Preinitiation Complex in Closed Conformation
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 4 of Structure of A Partial Yeast 48S Preinitiation Complex in Closed Conformation within 5.0Å range:
|
Magnesium binding site 5 out of 81 in 3jap
Go back to
Magnesium binding site 5 out
of 81 in the Structure of A Partial Yeast 48S Preinitiation Complex in Closed Conformation
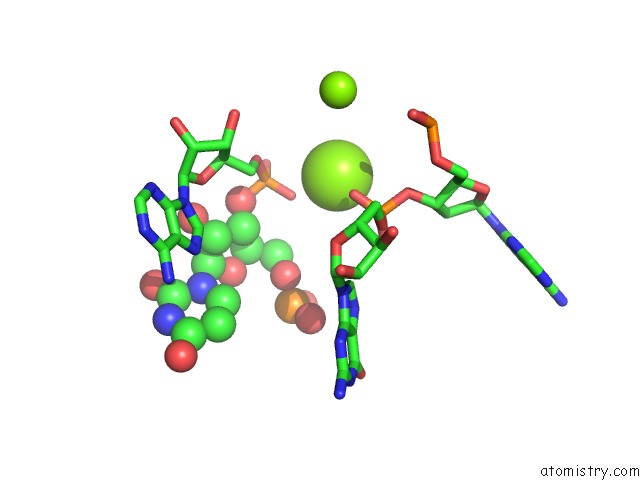
Mono view
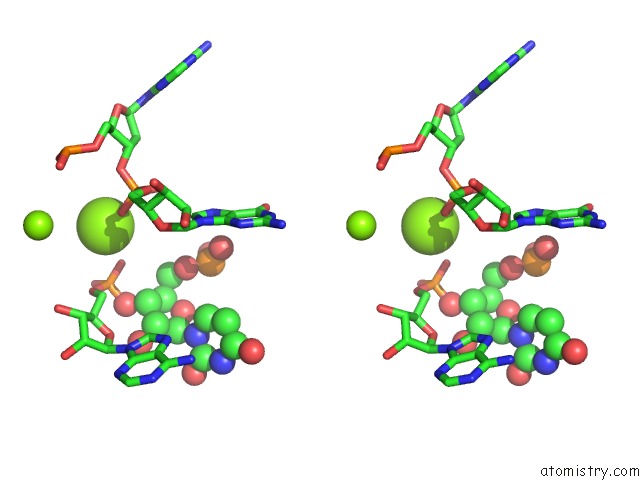
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 5 of Structure of A Partial Yeast 48S Preinitiation Complex in Closed Conformation within 5.0Å range:
|
Magnesium binding site 6 out of 81 in 3jap
Go back to
Magnesium binding site 6 out
of 81 in the Structure of A Partial Yeast 48S Preinitiation Complex in Closed Conformation
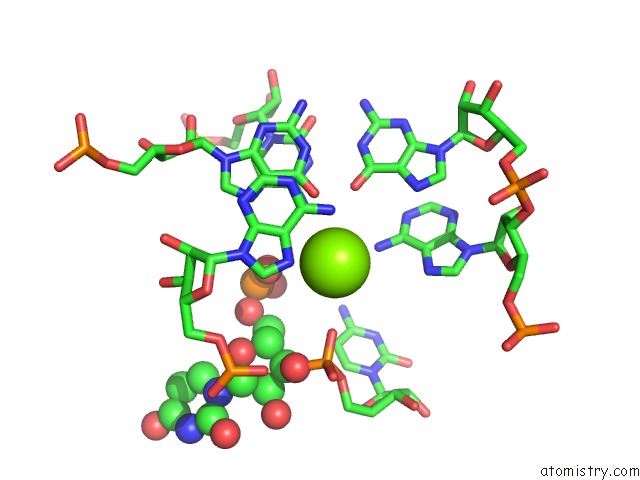
Mono view
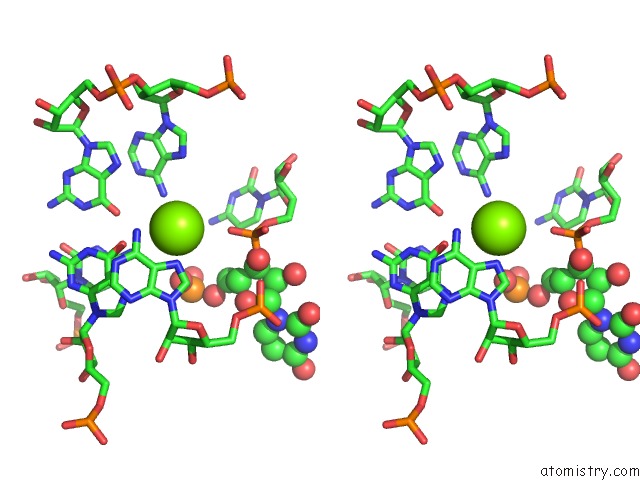
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 6 of Structure of A Partial Yeast 48S Preinitiation Complex in Closed Conformation within 5.0Å range:
|
Magnesium binding site 7 out of 81 in 3jap
Go back to
Magnesium binding site 7 out
of 81 in the Structure of A Partial Yeast 48S Preinitiation Complex in Closed Conformation
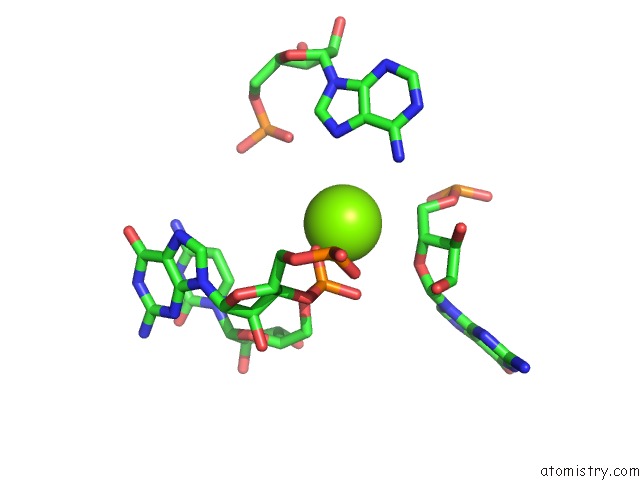
Mono view
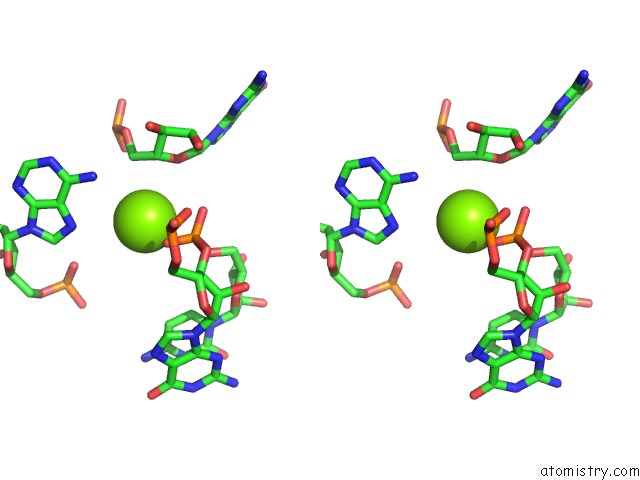
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 7 of Structure of A Partial Yeast 48S Preinitiation Complex in Closed Conformation within 5.0Å range:
|
Magnesium binding site 8 out of 81 in 3jap
Go back to
Magnesium binding site 8 out
of 81 in the Structure of A Partial Yeast 48S Preinitiation Complex in Closed Conformation
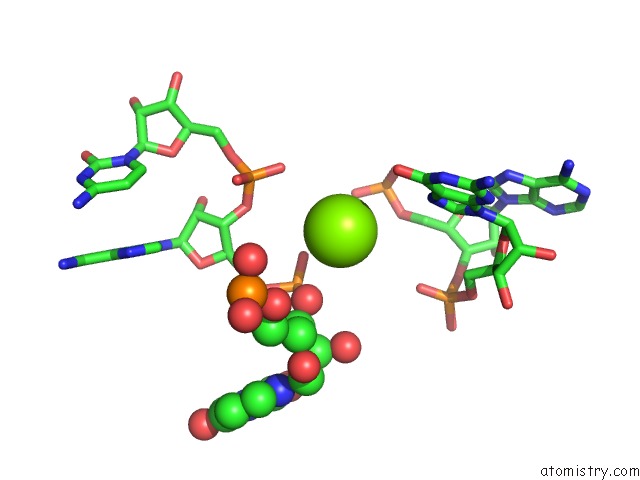
Mono view
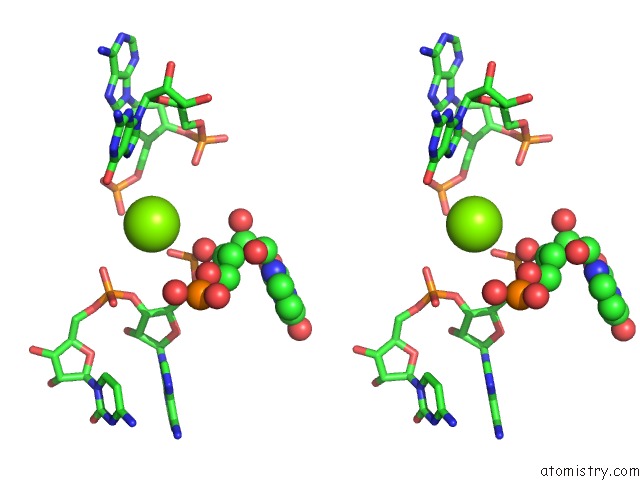
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 8 of Structure of A Partial Yeast 48S Preinitiation Complex in Closed Conformation within 5.0Å range:
|
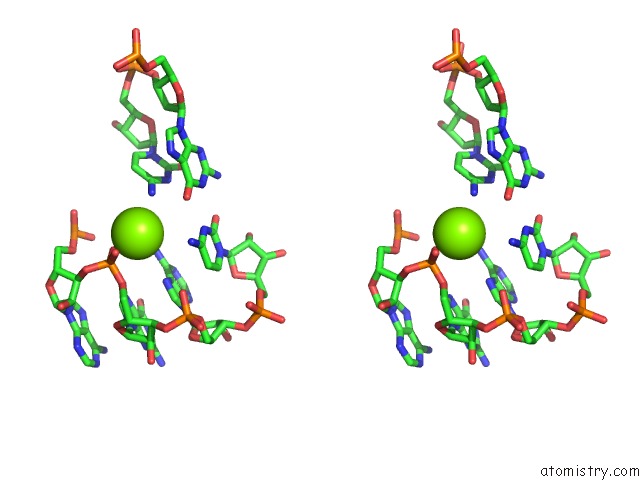
Magnesium binding site 9 out of 81 in 3jap
Go back to
Magnesium binding site 9 out
of 81 in the Structure of A Partial Yeast 48S Preinitiation Complex in Closed Conformation

Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 9 of Structure of A Partial Yeast 48S Preinitiation Complex in Closed Conformation within 5.0Å range:
|
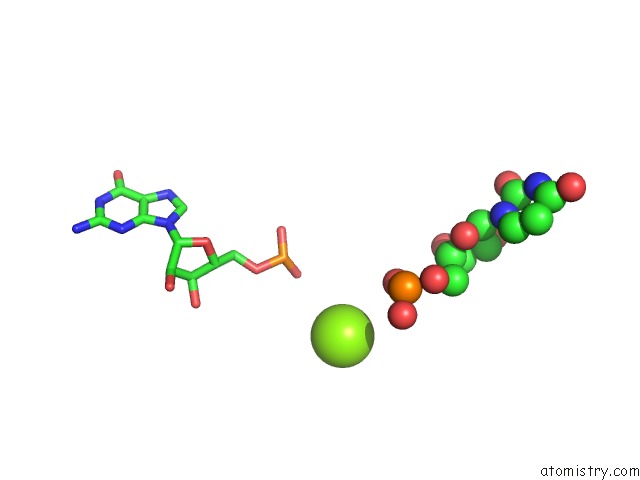
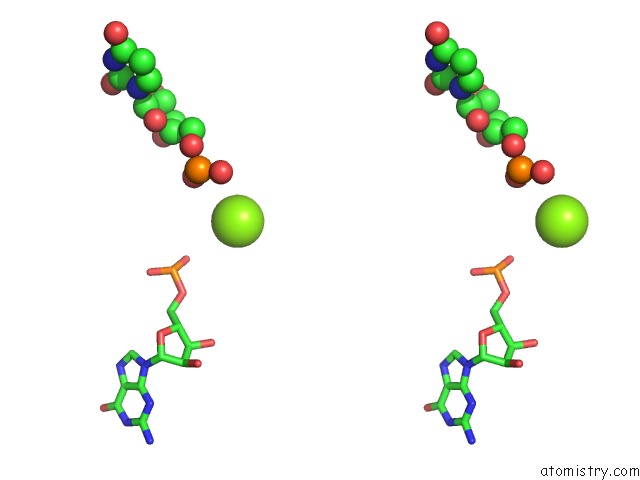
Magnesium binding site 10 out of 81 in 3jap
Go back to
Magnesium binding site 10 out
of 81 in the Structure of A Partial Yeast 48S Preinitiation Complex in Closed Conformation
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 10 of Structure of A Partial Yeast 48S Preinitiation Complex in Closed Conformation within 5.0Å range:
|
Reference:
J.L.Llacer,
T.Hussain,
L.Marler,
C.E.Aitken,
A.Thakur,
J.R.Lorsch,
A.G.Hinnebusch,
V.Ramakrishnan.
Conformational Differences Between Open and Closed States of the Eukaryotic Translation Initiation Complex. Mol.Cell V. 59 399 2015.
ISSN: ISSN 1097-2765
PubMed: 26212456
DOI: 10.1016/J.MOLCEL.2015.06.033
Page generated: Wed Aug 14 16:39:01 2024
ISSN: ISSN 1097-2765
PubMed: 26212456
DOI: 10.1016/J.MOLCEL.2015.06.033
Last articles
Fe in 2YXOFe in 2YRS
Fe in 2YXC
Fe in 2YNM
Fe in 2YVJ
Fe in 2YP1
Fe in 2YU2
Fe in 2YU1
Fe in 2YQB
Fe in 2YOO