Magnesium »
PDB 3ism-3jaw »
3jar »
Magnesium in PDB 3jar: Cryo-Em Structure of Gdp-Microtubule Co-Polymerized with EB3
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Cryo-Em Structure of Gdp-Microtubule Co-Polymerized with EB3
(pdb code 3jar). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 6 binding sites of Magnesium where determined in the Cryo-Em Structure of Gdp-Microtubule Co-Polymerized with EB3, PDB code: 3jar:
Jump to Magnesium binding site number: 1; 2; 3; 4; 5; 6;
In total 6 binding sites of Magnesium where determined in the Cryo-Em Structure of Gdp-Microtubule Co-Polymerized with EB3, PDB code: 3jar:
Jump to Magnesium binding site number: 1; 2; 3; 4; 5; 6;
Magnesium binding site 1 out of 6 in 3jar
Go back to
Magnesium binding site 1 out
of 6 in the Cryo-Em Structure of Gdp-Microtubule Co-Polymerized with EB3
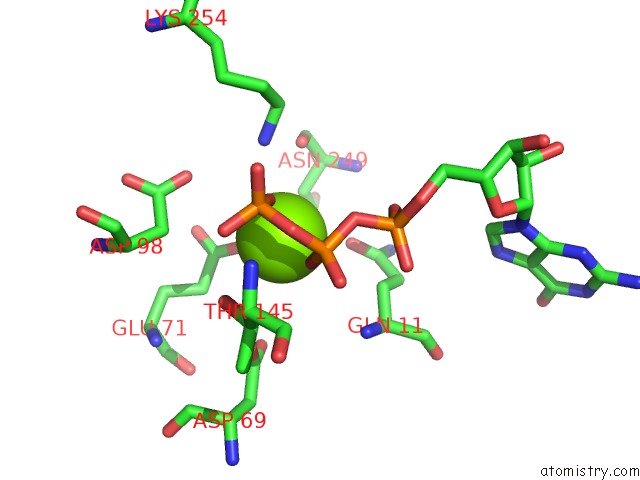
Mono view
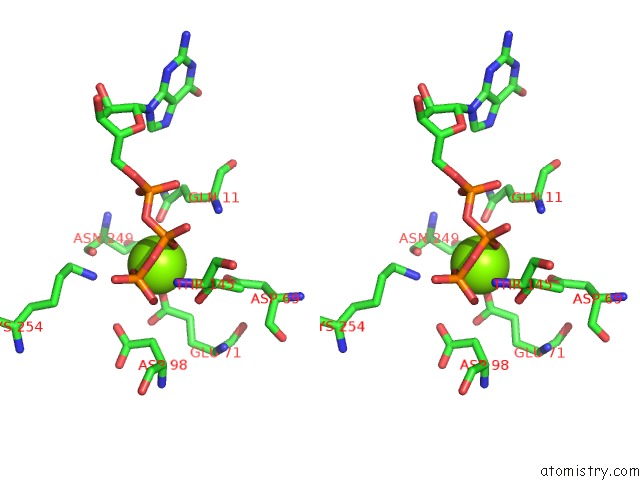
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Cryo-Em Structure of Gdp-Microtubule Co-Polymerized with EB3 within 5.0Å range:
|
Magnesium binding site 2 out of 6 in 3jar
Go back to
Magnesium binding site 2 out
of 6 in the Cryo-Em Structure of Gdp-Microtubule Co-Polymerized with EB3
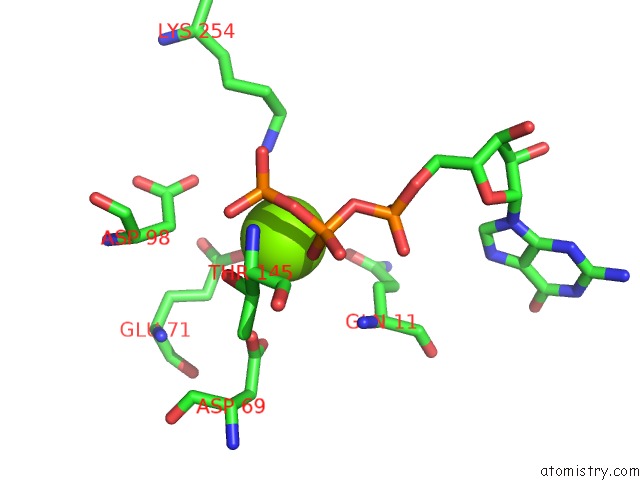
Mono view
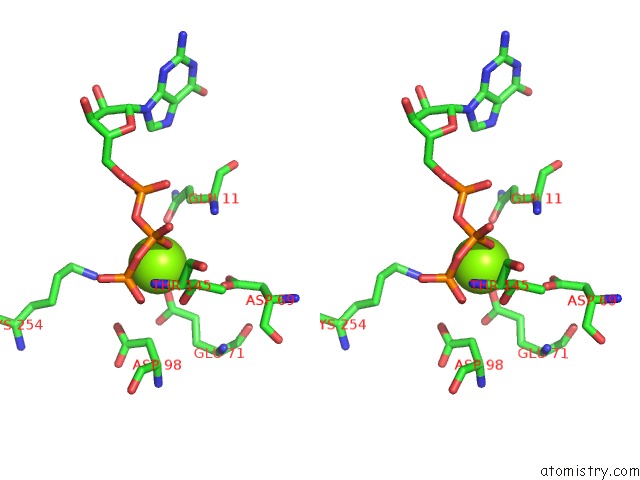
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Cryo-Em Structure of Gdp-Microtubule Co-Polymerized with EB3 within 5.0Å range:
|
Magnesium binding site 3 out of 6 in 3jar
Go back to
Magnesium binding site 3 out
of 6 in the Cryo-Em Structure of Gdp-Microtubule Co-Polymerized with EB3
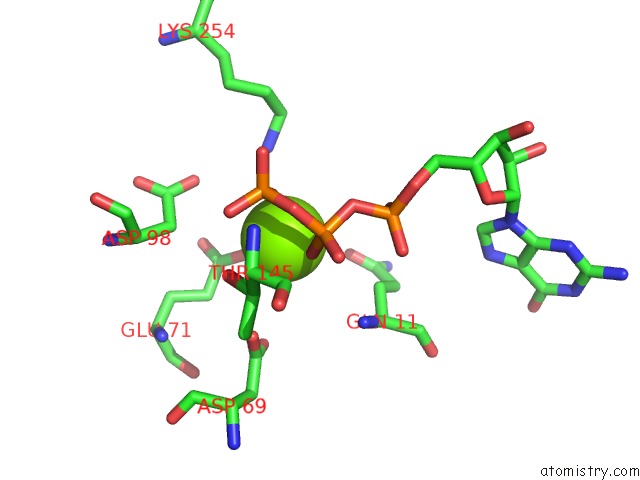
Mono view
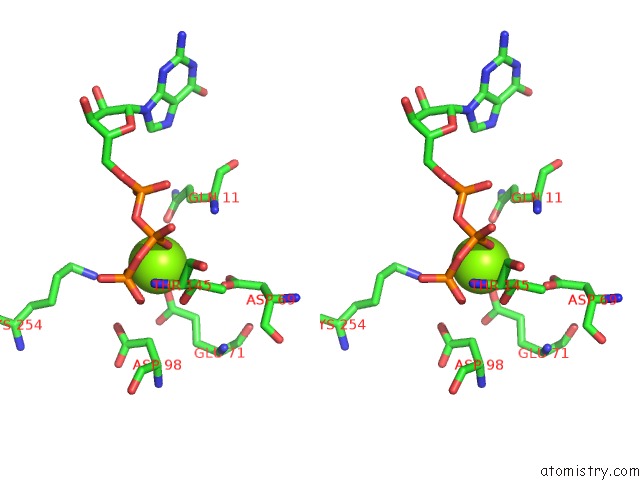
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 3 of Cryo-Em Structure of Gdp-Microtubule Co-Polymerized with EB3 within 5.0Å range:
|
Magnesium binding site 4 out of 6 in 3jar
Go back to
Magnesium binding site 4 out
of 6 in the Cryo-Em Structure of Gdp-Microtubule Co-Polymerized with EB3
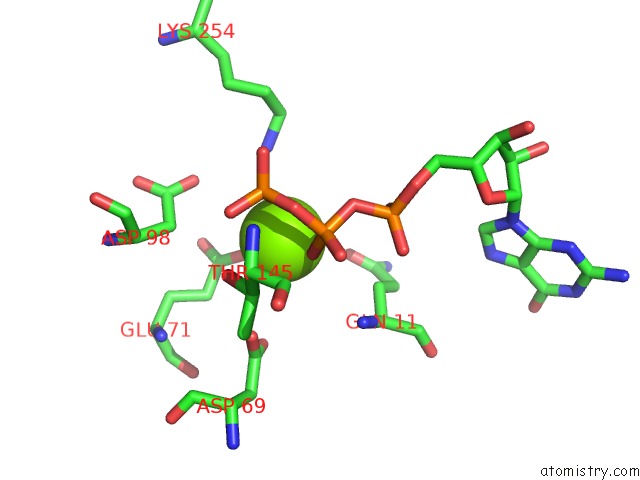
Mono view
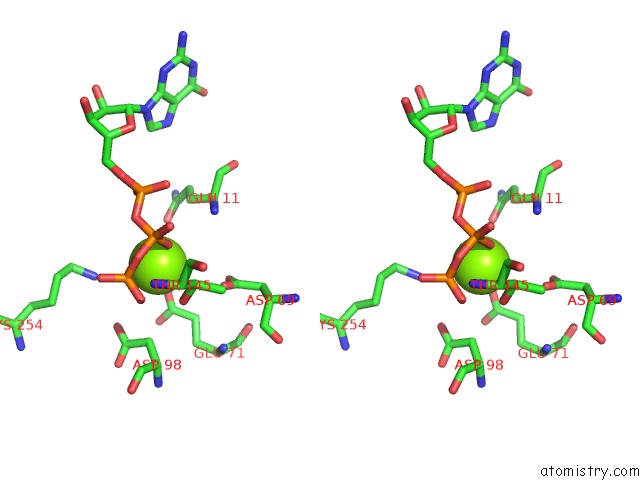
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 4 of Cryo-Em Structure of Gdp-Microtubule Co-Polymerized with EB3 within 5.0Å range:
|
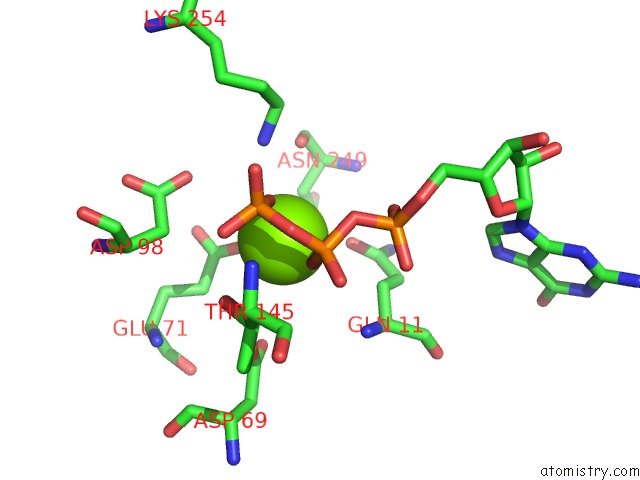
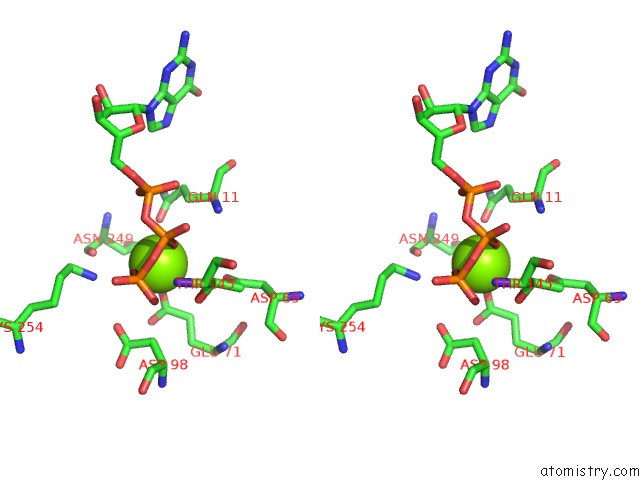
Magnesium binding site 5 out of 6 in 3jar
Go back to
Magnesium binding site 5 out
of 6 in the Cryo-Em Structure of Gdp-Microtubule Co-Polymerized with EB3
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 5 of Cryo-Em Structure of Gdp-Microtubule Co-Polymerized with EB3 within 5.0Å range:
|
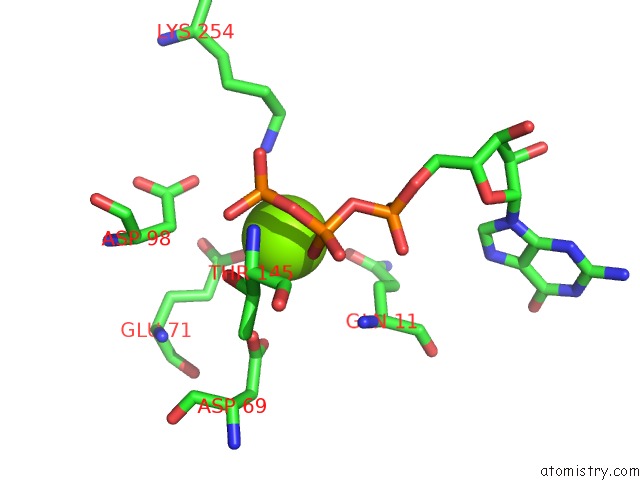
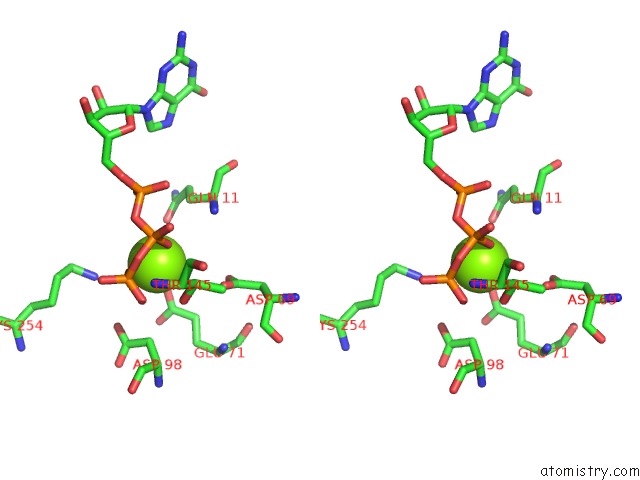
Magnesium binding site 6 out of 6 in 3jar
Go back to
Magnesium binding site 6 out
of 6 in the Cryo-Em Structure of Gdp-Microtubule Co-Polymerized with EB3
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 6 of Cryo-Em Structure of Gdp-Microtubule Co-Polymerized with EB3 within 5.0Å range:
|
Reference:
R.Zhang,
G.M.Alushin,
A.Brown,
E.Nogales.
Mechanistic Origin of Microtubule Dynamic Instability and Its Modulation By Eb Proteins. Cell(Cambridge,Mass.) V. 162 849 2015.
ISSN: ISSN 0092-8674
PubMed: 26234155
DOI: 10.1016/J.CELL.2015.07.012
Page generated: Wed Aug 14 16:58:13 2024
ISSN: ISSN 0092-8674
PubMed: 26234155
DOI: 10.1016/J.CELL.2015.07.012
Last articles
Zn in 9J0NZn in 9J0O
Zn in 9J0P
Zn in 9FJX
Zn in 9EKB
Zn in 9C0F
Zn in 9CAH
Zn in 9CH0
Zn in 9CH3
Zn in 9CH1