Magnesium »
PDB 3jxq-3k9f »
3k8d »
Magnesium in PDB 3k8d: Crystal Structure of E. Coli Lipopolysaccharide Specific Cmp-Kdo Synthetase in Complex with Ctp and 2-Deoxy-Kdo
Enzymatic activity of Crystal Structure of E. Coli Lipopolysaccharide Specific Cmp-Kdo Synthetase in Complex with Ctp and 2-Deoxy-Kdo
All present enzymatic activity of Crystal Structure of E. Coli Lipopolysaccharide Specific Cmp-Kdo Synthetase in Complex with Ctp and 2-Deoxy-Kdo:
2.7.7.38;
2.7.7.38;
Protein crystallography data
The structure of Crystal Structure of E. Coli Lipopolysaccharide Specific Cmp-Kdo Synthetase in Complex with Ctp and 2-Deoxy-Kdo, PDB code: 3k8d
was solved by
D.J.Heyes,
C.W.Levy,
P.Lafite,
N.S.Scrutton,
D.Leys,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 45.27 / 1.90 |
| Space group | P 31 |
| Cell size a, b, c (Å), α, β, γ (°) | 94.770, 94.770, 153.140, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 15.1 / 18.2 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Crystal Structure of E. Coli Lipopolysaccharide Specific Cmp-Kdo Synthetase in Complex with Ctp and 2-Deoxy-Kdo
(pdb code 3k8d). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 4 binding sites of Magnesium where determined in the Crystal Structure of E. Coli Lipopolysaccharide Specific Cmp-Kdo Synthetase in Complex with Ctp and 2-Deoxy-Kdo, PDB code: 3k8d:
Jump to Magnesium binding site number: 1; 2; 3; 4;
In total 4 binding sites of Magnesium where determined in the Crystal Structure of E. Coli Lipopolysaccharide Specific Cmp-Kdo Synthetase in Complex with Ctp and 2-Deoxy-Kdo, PDB code: 3k8d:
Jump to Magnesium binding site number: 1; 2; 3; 4;
Magnesium binding site 1 out of 4 in 3k8d
Go back to
Magnesium binding site 1 out
of 4 in the Crystal Structure of E. Coli Lipopolysaccharide Specific Cmp-Kdo Synthetase in Complex with Ctp and 2-Deoxy-Kdo
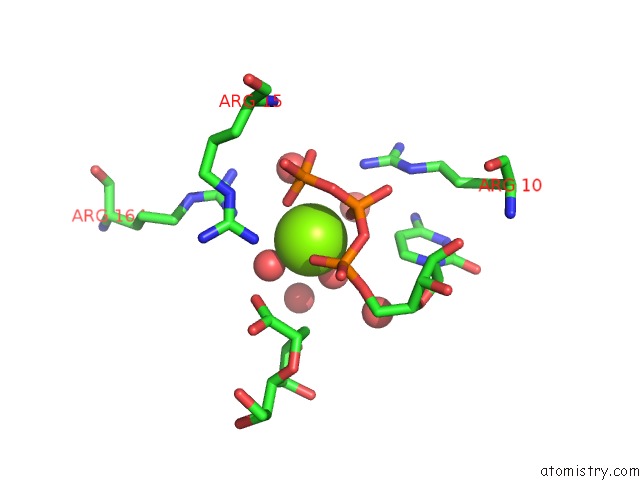
Mono view
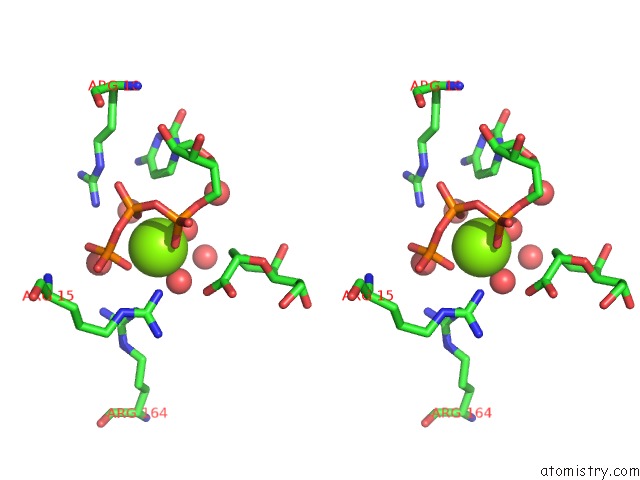
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Crystal Structure of E. Coli Lipopolysaccharide Specific Cmp-Kdo Synthetase in Complex with Ctp and 2-Deoxy-Kdo within 5.0Å range:
|
Magnesium binding site 2 out of 4 in 3k8d
Go back to
Magnesium binding site 2 out
of 4 in the Crystal Structure of E. Coli Lipopolysaccharide Specific Cmp-Kdo Synthetase in Complex with Ctp and 2-Deoxy-Kdo
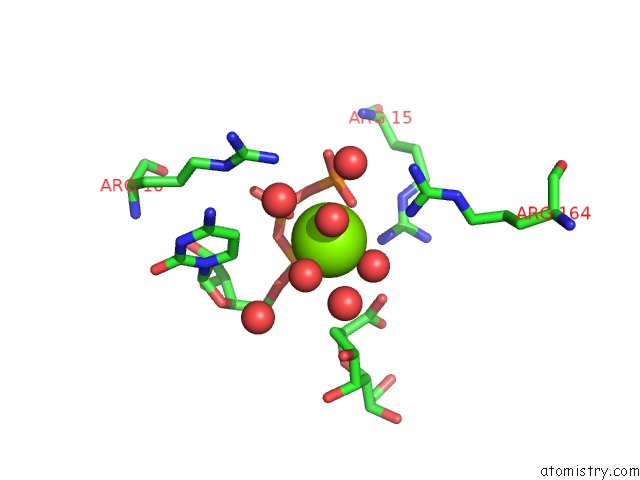
Mono view
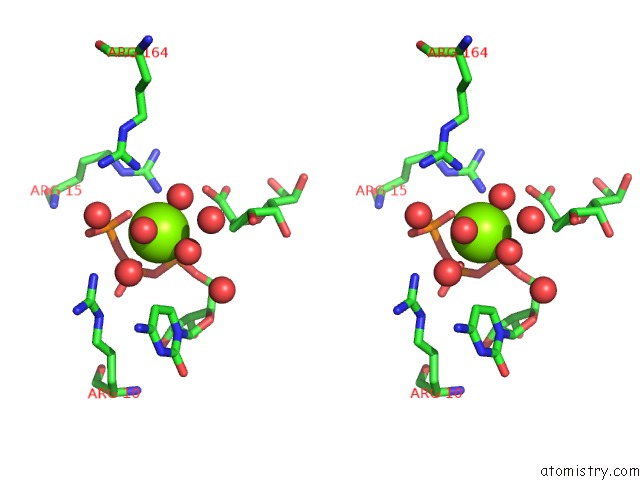
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Crystal Structure of E. Coli Lipopolysaccharide Specific Cmp-Kdo Synthetase in Complex with Ctp and 2-Deoxy-Kdo within 5.0Å range:
|
Magnesium binding site 3 out of 4 in 3k8d
Go back to
Magnesium binding site 3 out
of 4 in the Crystal Structure of E. Coli Lipopolysaccharide Specific Cmp-Kdo Synthetase in Complex with Ctp and 2-Deoxy-Kdo
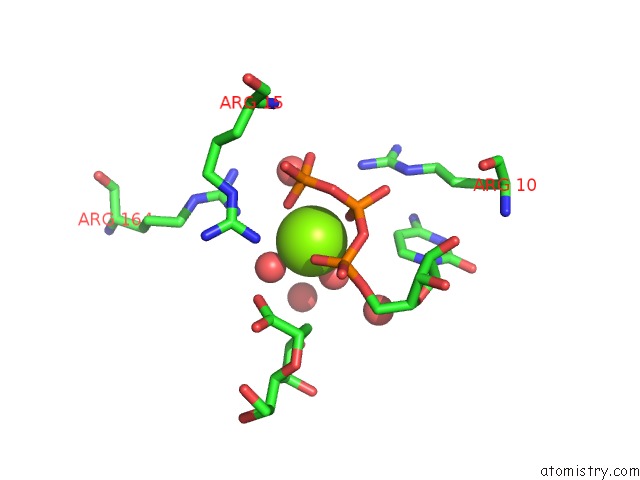
Mono view
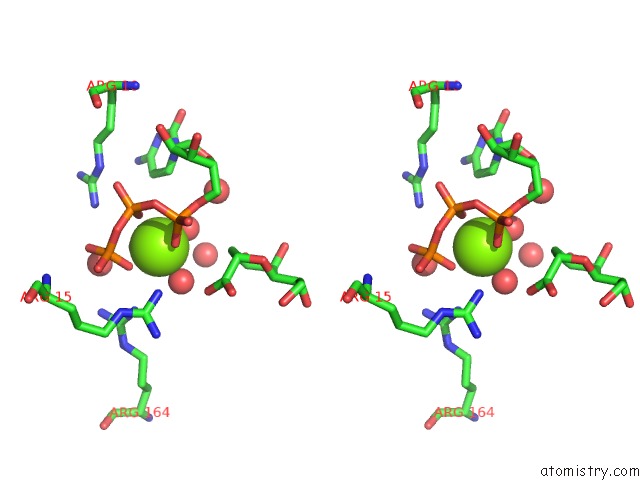
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 3 of Crystal Structure of E. Coli Lipopolysaccharide Specific Cmp-Kdo Synthetase in Complex with Ctp and 2-Deoxy-Kdo within 5.0Å range:
|
Magnesium binding site 4 out of 4 in 3k8d
Go back to
Magnesium binding site 4 out
of 4 in the Crystal Structure of E. Coli Lipopolysaccharide Specific Cmp-Kdo Synthetase in Complex with Ctp and 2-Deoxy-Kdo
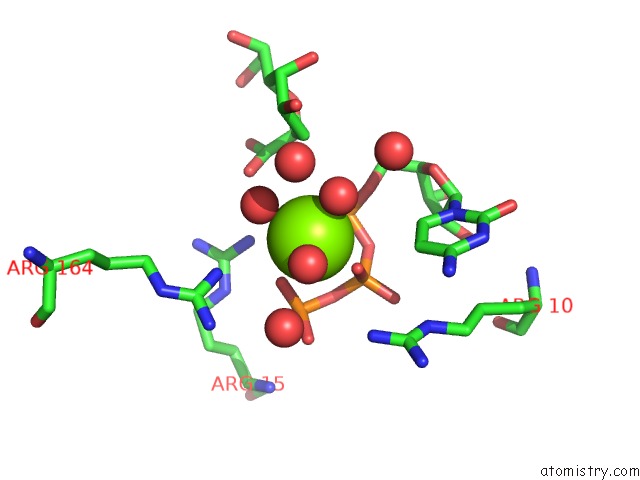
Mono view
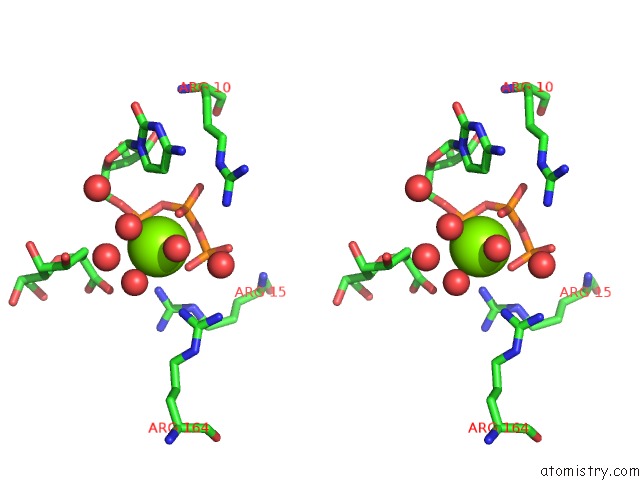
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 4 of Crystal Structure of E. Coli Lipopolysaccharide Specific Cmp-Kdo Synthetase in Complex with Ctp and 2-Deoxy-Kdo within 5.0Å range:
|
Reference:
D.J.Heyes,
C.Levy,
P.Lafite,
I.S.Roberts,
M.Goldrick,
A.V.Stachulski,
S.B.Rossington,
D.Stanford,
S.E.J.Rigby,
N.S.Scrutton,
D.Leys.
Structure-Based Mechanism of Cmp-2-Keto-3-Deoxymanno-Octulonic Acid Synthetase: Convergent Evolution of A Sugar-Activating Enzyme with Dna/Rna Polymerases J.Biol.Chem. V. 284 35514 2009.
ISSN: ISSN 0021-9258
PubMed: 19815542
DOI: 10.1074/JBC.M109.056630
Page generated: Wed Aug 14 17:57:02 2024
ISSN: ISSN 0021-9258
PubMed: 19815542
DOI: 10.1074/JBC.M109.056630
Last articles
Cl in 7UVOCl in 7UVF
Cl in 7UQC
Cl in 7UQA
Cl in 7UUP
Cl in 7UUL
Cl in 7UUK
Cl in 7USI
Cl in 7UUJ
Cl in 7UUG