Magnesium »
PDB 3l9b-3lop »
3ld0 »
Magnesium in PDB 3ld0: Crystal Structure of B.Licheniformis Anti-Trap Protein, An Antagonist of Trap-Rna Interactions
Protein crystallography data
The structure of Crystal Structure of B.Licheniformis Anti-Trap Protein, An Antagonist of Trap-Rna Interactions, PDB code: 3ld0
was solved by
M.B.Shevtsov,
Y.Chen,
M.N.Isupov,
P.Gollnick,
A.A.Antson,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 29.27 / 2.20 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 118.544, 99.865, 123.178, 90.00, 117.61, 90.00 |
| R / Rfree (%) | 19.4 / 25.9 |
Other elements in 3ld0:
The structure of Crystal Structure of B.Licheniformis Anti-Trap Protein, An Antagonist of Trap-Rna Interactions also contains other interesting chemical elements:
| Zinc | (Zn) | 48 atoms |
Magnesium Binding Sites:
Pages:
>>> Page 1 <<< Page 2, Binding sites: 11 - 16;Binding sites:
The binding sites of Magnesium atom in the Crystal Structure of B.Licheniformis Anti-Trap Protein, An Antagonist of Trap-Rna Interactions (pdb code 3ld0). This binding sites where shown within 5.0 Angstroms radius around Magnesium atom.In total 16 binding sites of Magnesium where determined in the Crystal Structure of B.Licheniformis Anti-Trap Protein, An Antagonist of Trap-Rna Interactions, PDB code: 3ld0:
Jump to Magnesium binding site number: 1; 2; 3; 4; 5; 6; 7; 8; 9; 10;
Magnesium binding site 1 out of 16 in 3ld0
Go back to
Magnesium binding site 1 out
of 16 in the Crystal Structure of B.Licheniformis Anti-Trap Protein, An Antagonist of Trap-Rna Interactions
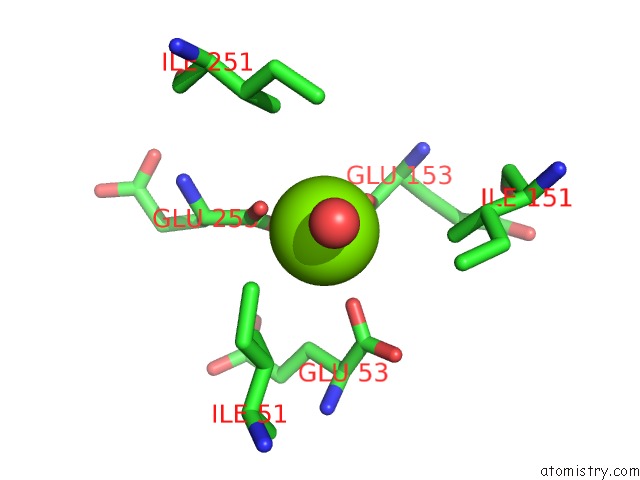
Mono view
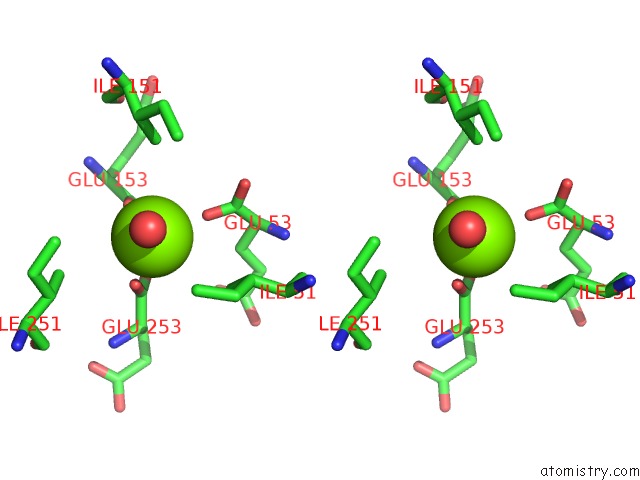
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Crystal Structure of B.Licheniformis Anti-Trap Protein, An Antagonist of Trap-Rna Interactions within 5.0Å range:
|
Magnesium binding site 2 out of 16 in 3ld0
Go back to
Magnesium binding site 2 out
of 16 in the Crystal Structure of B.Licheniformis Anti-Trap Protein, An Antagonist of Trap-Rna Interactions

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Crystal Structure of B.Licheniformis Anti-Trap Protein, An Antagonist of Trap-Rna Interactions within 5.0Å range:
|
Magnesium binding site 3 out of 16 in 3ld0
Go back to
Magnesium binding site 3 out
of 16 in the Crystal Structure of B.Licheniformis Anti-Trap Protein, An Antagonist of Trap-Rna Interactions

Mono view
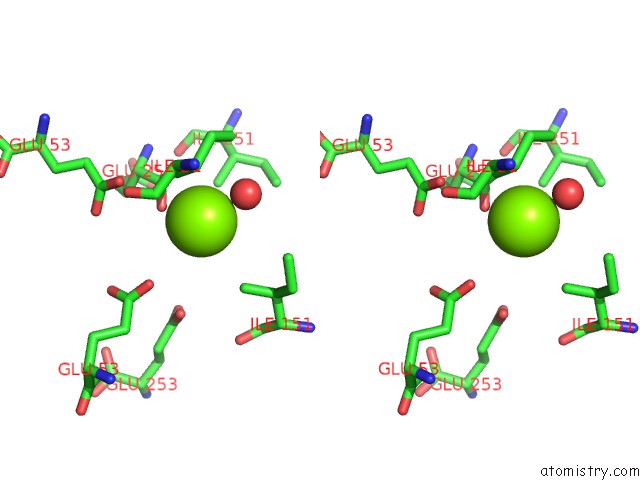
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 3 of Crystal Structure of B.Licheniformis Anti-Trap Protein, An Antagonist of Trap-Rna Interactions within 5.0Å range:
|
Magnesium binding site 4 out of 16 in 3ld0
Go back to
Magnesium binding site 4 out
of 16 in the Crystal Structure of B.Licheniformis Anti-Trap Protein, An Antagonist of Trap-Rna Interactions
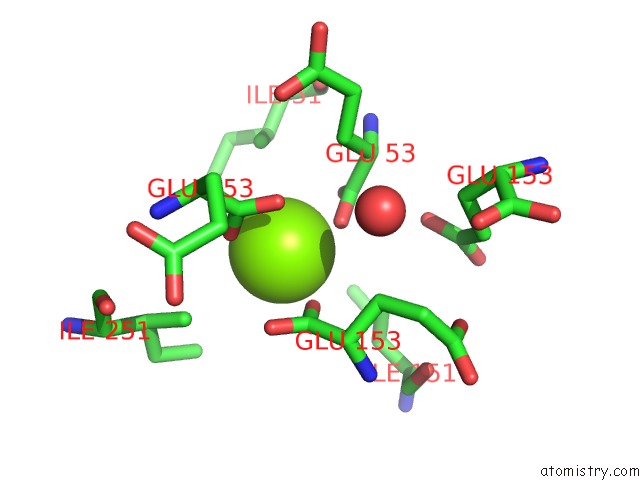
Mono view
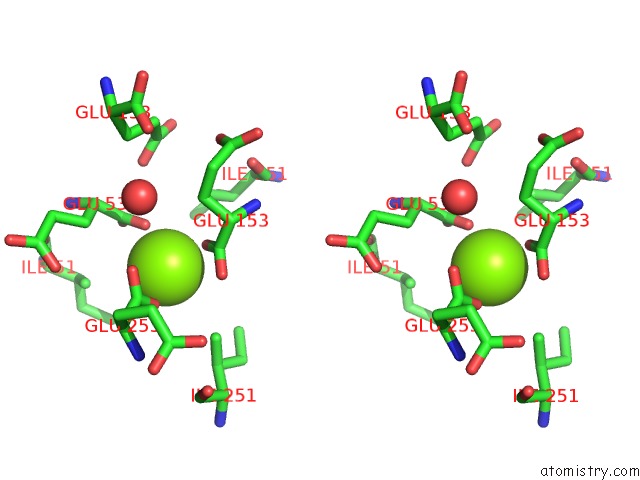
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 4 of Crystal Structure of B.Licheniformis Anti-Trap Protein, An Antagonist of Trap-Rna Interactions within 5.0Å range:
|
Magnesium binding site 5 out of 16 in 3ld0
Go back to
Magnesium binding site 5 out
of 16 in the Crystal Structure of B.Licheniformis Anti-Trap Protein, An Antagonist of Trap-Rna Interactions

Mono view
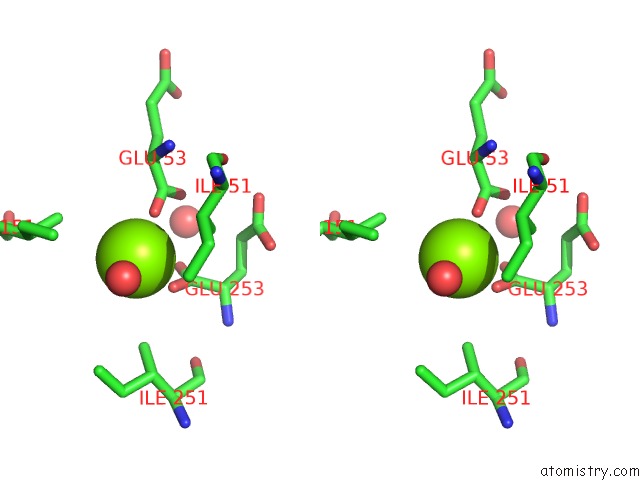
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 5 of Crystal Structure of B.Licheniformis Anti-Trap Protein, An Antagonist of Trap-Rna Interactions within 5.0Å range:
|
Magnesium binding site 6 out of 16 in 3ld0
Go back to
Magnesium binding site 6 out
of 16 in the Crystal Structure of B.Licheniformis Anti-Trap Protein, An Antagonist of Trap-Rna Interactions
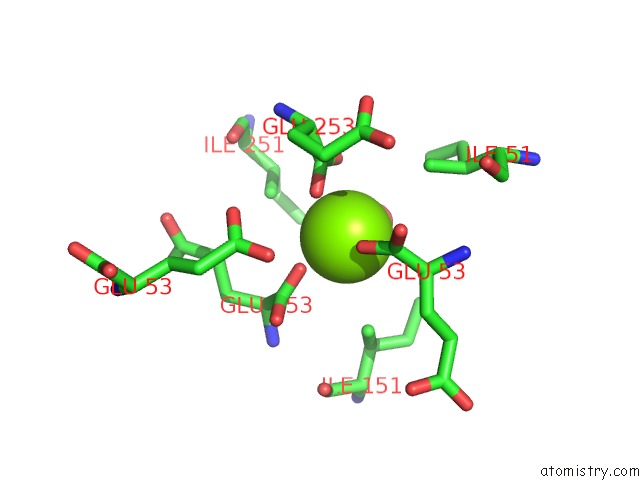
Mono view
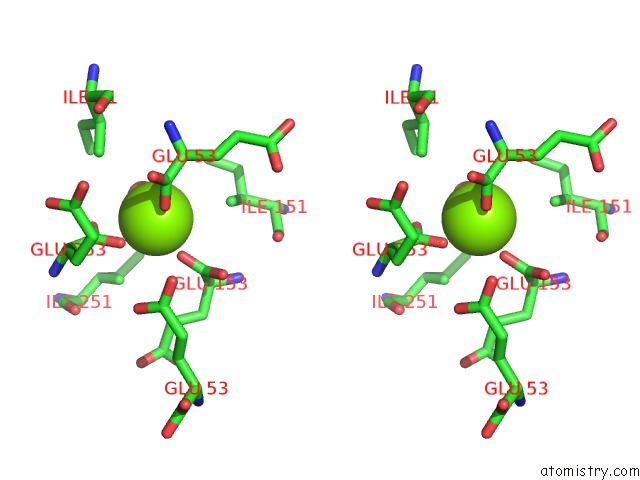
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 6 of Crystal Structure of B.Licheniformis Anti-Trap Protein, An Antagonist of Trap-Rna Interactions within 5.0Å range:
|
Magnesium binding site 7 out of 16 in 3ld0
Go back to
Magnesium binding site 7 out
of 16 in the Crystal Structure of B.Licheniformis Anti-Trap Protein, An Antagonist of Trap-Rna Interactions
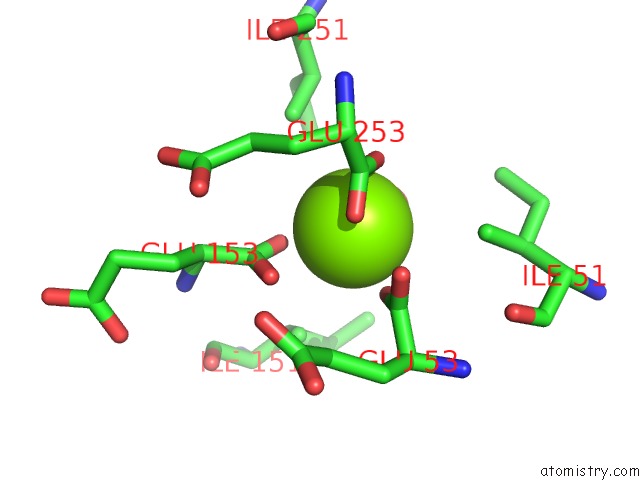
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 7 of Crystal Structure of B.Licheniformis Anti-Trap Protein, An Antagonist of Trap-Rna Interactions within 5.0Å range:
|
Magnesium binding site 8 out of 16 in 3ld0
Go back to
Magnesium binding site 8 out
of 16 in the Crystal Structure of B.Licheniformis Anti-Trap Protein, An Antagonist of Trap-Rna Interactions
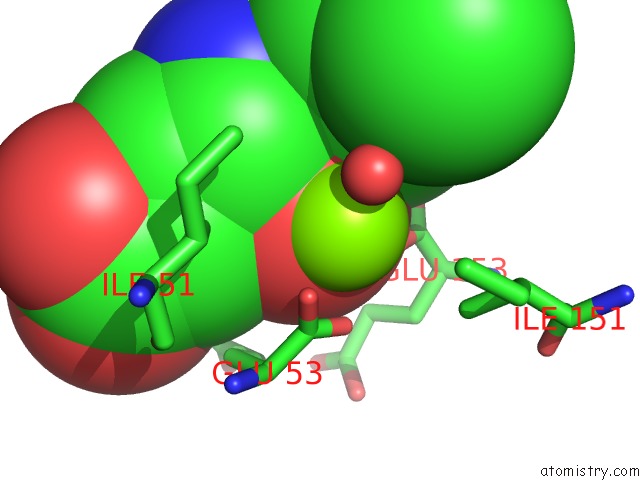
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 8 of Crystal Structure of B.Licheniformis Anti-Trap Protein, An Antagonist of Trap-Rna Interactions within 5.0Å range:
|
Magnesium binding site 9 out of 16 in 3ld0
Go back to
Magnesium binding site 9 out
of 16 in the Crystal Structure of B.Licheniformis Anti-Trap Protein, An Antagonist of Trap-Rna Interactions
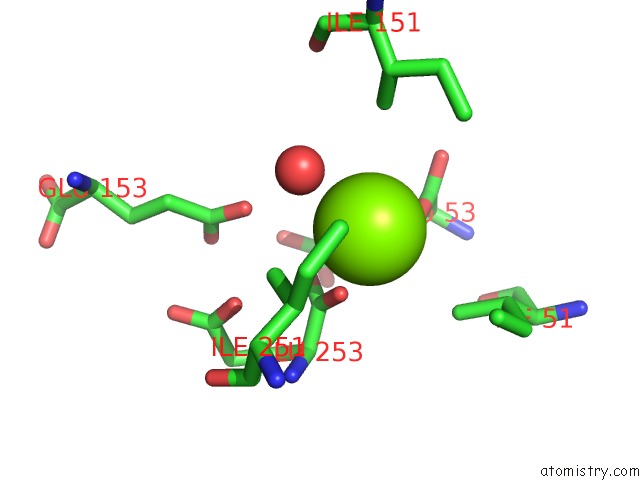
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 9 of Crystal Structure of B.Licheniformis Anti-Trap Protein, An Antagonist of Trap-Rna Interactions within 5.0Å range:
|
Magnesium binding site 10 out of 16 in 3ld0
Go back to
Magnesium binding site 10 out
of 16 in the Crystal Structure of B.Licheniformis Anti-Trap Protein, An Antagonist of Trap-Rna Interactions

Mono view
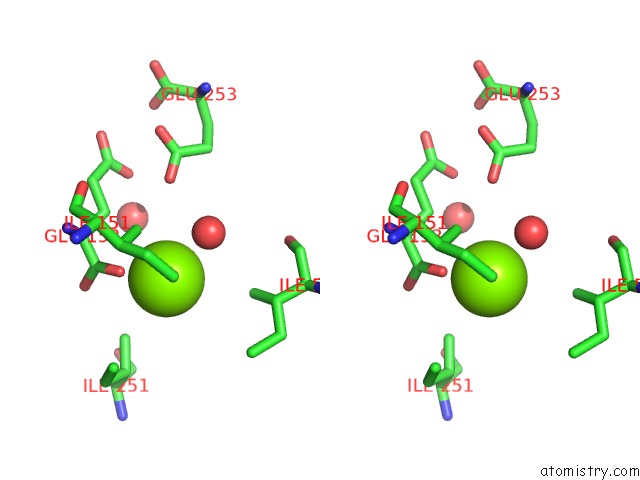
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 10 of Crystal Structure of B.Licheniformis Anti-Trap Protein, An Antagonist of Trap-Rna Interactions within 5.0Å range:
|
Reference:
M.B.Shevtsov,
Y.Chen,
M.N.Isupov,
A.Leech,
P.Gollnick,
A.A.Antson.
Bacillus Licheniformis Anti-Trap Can Assemble Into Two Types of Dodecameric Particles with the Same Symmetry But Inverted Orientation of Trimers. J.Struct.Biol. V. 170 127 2010.
ISSN: ISSN 1047-8477
PubMed: 20138150
DOI: 10.1016/J.JSB.2010.01.013
Page generated: Wed Aug 14 18:30:40 2024
ISSN: ISSN 1047-8477
PubMed: 20138150
DOI: 10.1016/J.JSB.2010.01.013
Last articles
F in 7K1FF in 7K1E
F in 7K1I
F in 7K1D
F in 7JY4
F in 7K0V
F in 7JYM
F in 7K15
F in 7K12
F in 7K0W