Magnesium »
PDB 3q89-3qpo »
3qlz »
Magnesium in PDB 3qlz: Candida Glabrata Dihydrofolate Reductase Complexed with Nadph and 5- [3-(2,5-Dimethoxyphenyl)Prop-1-Yn-1-Yl]-6-Propylpyrimidine-2,4- Diamine (UCP130B)
Protein crystallography data
The structure of Candida Glabrata Dihydrofolate Reductase Complexed with Nadph and 5- [3-(2,5-Dimethoxyphenyl)Prop-1-Yn-1-Yl]-6-Propylpyrimidine-2,4- Diamine (UCP130B), PDB code: 3qlz
was solved by
J.L.Paulsen,
S.D.Bendel,
A.C.Anderson,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 28.78 / 1.94 |
| Space group | P 41 |
| Cell size a, b, c (Å), α, β, γ (°) | 42.607, 42.607, 230.236, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 20.6 / 24.5 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Candida Glabrata Dihydrofolate Reductase Complexed with Nadph and 5- [3-(2,5-Dimethoxyphenyl)Prop-1-Yn-1-Yl]-6-Propylpyrimidine-2,4- Diamine (UCP130B)
(pdb code 3qlz). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 3 binding sites of Magnesium where determined in the Candida Glabrata Dihydrofolate Reductase Complexed with Nadph and 5- [3-(2,5-Dimethoxyphenyl)Prop-1-Yn-1-Yl]-6-Propylpyrimidine-2,4- Diamine (UCP130B), PDB code: 3qlz:
Jump to Magnesium binding site number: 1; 2; 3;
In total 3 binding sites of Magnesium where determined in the Candida Glabrata Dihydrofolate Reductase Complexed with Nadph and 5- [3-(2,5-Dimethoxyphenyl)Prop-1-Yn-1-Yl]-6-Propylpyrimidine-2,4- Diamine (UCP130B), PDB code: 3qlz:
Jump to Magnesium binding site number: 1; 2; 3;
Magnesium binding site 1 out of 3 in 3qlz
Go back to
Magnesium binding site 1 out
of 3 in the Candida Glabrata Dihydrofolate Reductase Complexed with Nadph and 5- [3-(2,5-Dimethoxyphenyl)Prop-1-Yn-1-Yl]-6-Propylpyrimidine-2,4- Diamine (UCP130B)
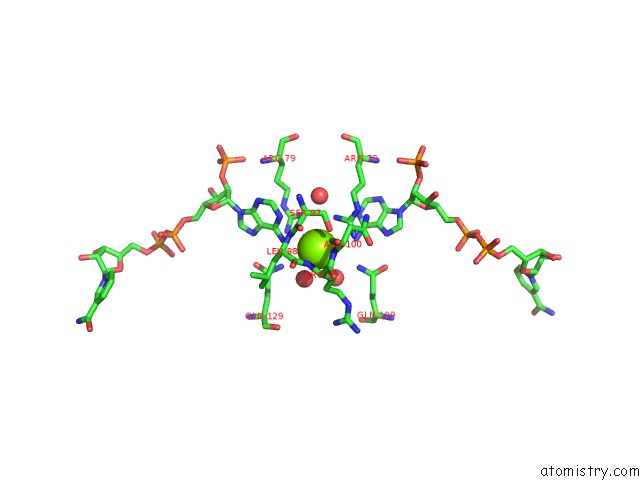
Mono view
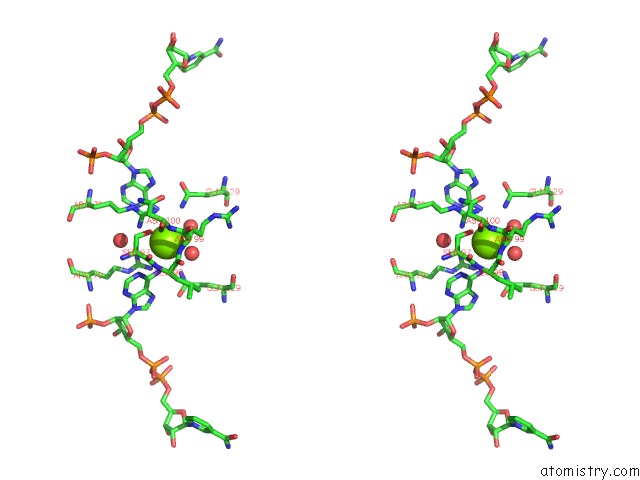
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Candida Glabrata Dihydrofolate Reductase Complexed with Nadph and 5- [3-(2,5-Dimethoxyphenyl)Prop-1-Yn-1-Yl]-6-Propylpyrimidine-2,4- Diamine (UCP130B) within 5.0Å range:
|
Magnesium binding site 2 out of 3 in 3qlz
Go back to
Magnesium binding site 2 out
of 3 in the Candida Glabrata Dihydrofolate Reductase Complexed with Nadph and 5- [3-(2,5-Dimethoxyphenyl)Prop-1-Yn-1-Yl]-6-Propylpyrimidine-2,4- Diamine (UCP130B)
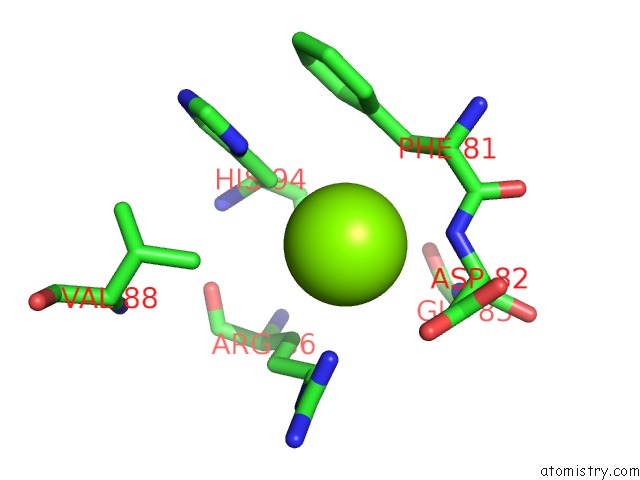
Mono view
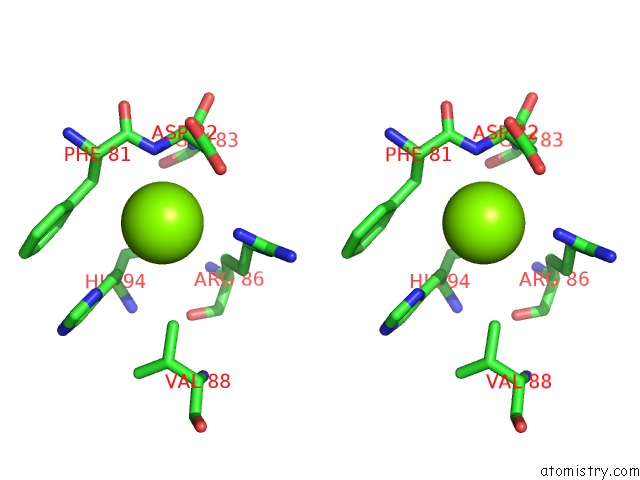
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Candida Glabrata Dihydrofolate Reductase Complexed with Nadph and 5- [3-(2,5-Dimethoxyphenyl)Prop-1-Yn-1-Yl]-6-Propylpyrimidine-2,4- Diamine (UCP130B) within 5.0Å range:
|
Magnesium binding site 3 out of 3 in 3qlz
Go back to
Magnesium binding site 3 out
of 3 in the Candida Glabrata Dihydrofolate Reductase Complexed with Nadph and 5- [3-(2,5-Dimethoxyphenyl)Prop-1-Yn-1-Yl]-6-Propylpyrimidine-2,4- Diamine (UCP130B)

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 3 of Candida Glabrata Dihydrofolate Reductase Complexed with Nadph and 5- [3-(2,5-Dimethoxyphenyl)Prop-1-Yn-1-Yl]-6-Propylpyrimidine-2,4- Diamine (UCP130B) within 5.0Å range:
|
Reference:
J.L.Paulsen,
S.D.Bendel,
A.C.Anderson.
Crystal Structures of Candida Albicans Dihydrofolate Reductase Bound to Propargyl-Linked Antifolates Reveal the Flexibility of Active Site Loop Residues Critical For Ligand Potency and Selectivity. Chem.Biol.Drug Des. V. 78 505 2011.
ISSN: ISSN 1747-0277
PubMed: 21726415
DOI: 10.1111/J.1747-0285.2011.01169.X
Page generated: Thu Aug 15 10:04:45 2024
ISSN: ISSN 1747-0277
PubMed: 21726415
DOI: 10.1111/J.1747-0285.2011.01169.X
Last articles
F in 7JL0F in 7JH2
F in 7JK8
F in 7JHW
F in 7JHD
F in 7I18
F in 7I2F
F in 7I2M
F in 7I2A
F in 7I2D