Magnesium »
PDB 3rvb-3s8c »
3s1a »
Magnesium in PDB 3s1a: Crystal Structure of the Phosphorylation-Site Double Mutant S431E/T432E of the Kaic Circadian Clock Protein
Enzymatic activity of Crystal Structure of the Phosphorylation-Site Double Mutant S431E/T432E of the Kaic Circadian Clock Protein
All present enzymatic activity of Crystal Structure of the Phosphorylation-Site Double Mutant S431E/T432E of the Kaic Circadian Clock Protein:
2.7.11.1;
2.7.11.1;
Protein crystallography data
The structure of Crystal Structure of the Phosphorylation-Site Double Mutant S431E/T432E of the Kaic Circadian Clock Protein, PDB code: 3s1a
was solved by
R.Pattanayek,
D.W.Williams,
G.Rossi,
S.Weigand,
T.Mori,
C.H.Johnson,
P.L.Stewart,
M.Egli,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 17.00 / 3.00 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 132.666, 135.494, 204.603, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 24.2 / 28.8 |
Magnesium Binding Sites:
Pages:
>>> Page 1 <<< Page 2, Binding sites: 11 - 20; Page 3, Binding sites: 21 - 21;Binding sites:
The binding sites of Magnesium atom in the Crystal Structure of the Phosphorylation-Site Double Mutant S431E/T432E of the Kaic Circadian Clock Protein (pdb code 3s1a). This binding sites where shown within 5.0 Angstroms radius around Magnesium atom.In total 21 binding sites of Magnesium where determined in the Crystal Structure of the Phosphorylation-Site Double Mutant S431E/T432E of the Kaic Circadian Clock Protein, PDB code: 3s1a:
Jump to Magnesium binding site number: 1; 2; 3; 4; 5; 6; 7; 8; 9; 10;
Magnesium binding site 1 out of 21 in 3s1a
Go back to
Magnesium binding site 1 out
of 21 in the Crystal Structure of the Phosphorylation-Site Double Mutant S431E/T432E of the Kaic Circadian Clock Protein

Mono view
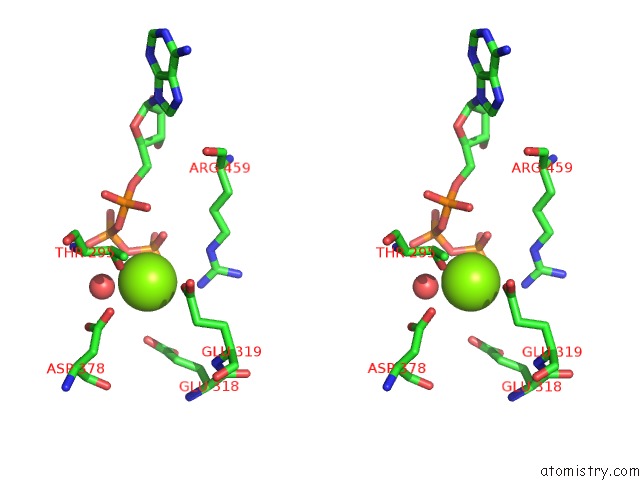
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Crystal Structure of the Phosphorylation-Site Double Mutant S431E/T432E of the Kaic Circadian Clock Protein within 5.0Å range:
|
Magnesium binding site 2 out of 21 in 3s1a
Go back to
Magnesium binding site 2 out
of 21 in the Crystal Structure of the Phosphorylation-Site Double Mutant S431E/T432E of the Kaic Circadian Clock Protein
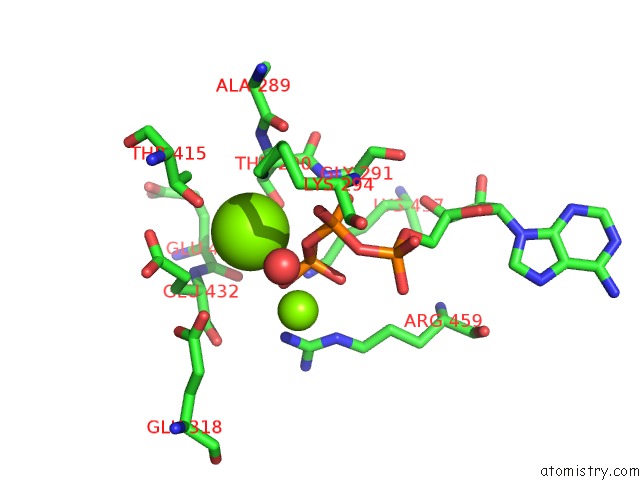
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Crystal Structure of the Phosphorylation-Site Double Mutant S431E/T432E of the Kaic Circadian Clock Protein within 5.0Å range:
|
Magnesium binding site 3 out of 21 in 3s1a
Go back to
Magnesium binding site 3 out
of 21 in the Crystal Structure of the Phosphorylation-Site Double Mutant S431E/T432E of the Kaic Circadian Clock Protein

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 3 of Crystal Structure of the Phosphorylation-Site Double Mutant S431E/T432E of the Kaic Circadian Clock Protein within 5.0Å range:
|
Magnesium binding site 4 out of 21 in 3s1a
Go back to
Magnesium binding site 4 out
of 21 in the Crystal Structure of the Phosphorylation-Site Double Mutant S431E/T432E of the Kaic Circadian Clock Protein
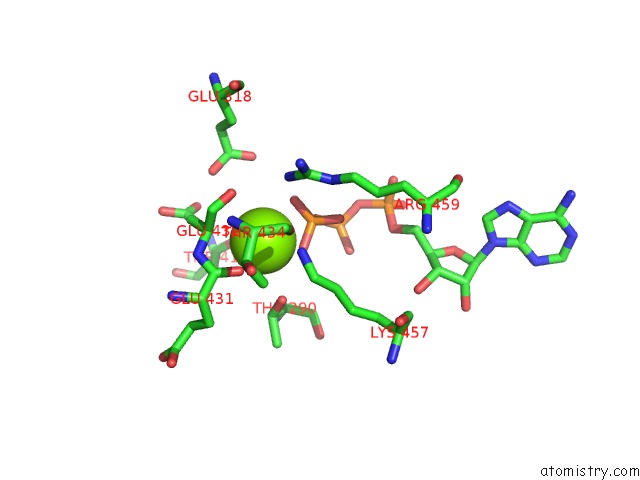
Mono view
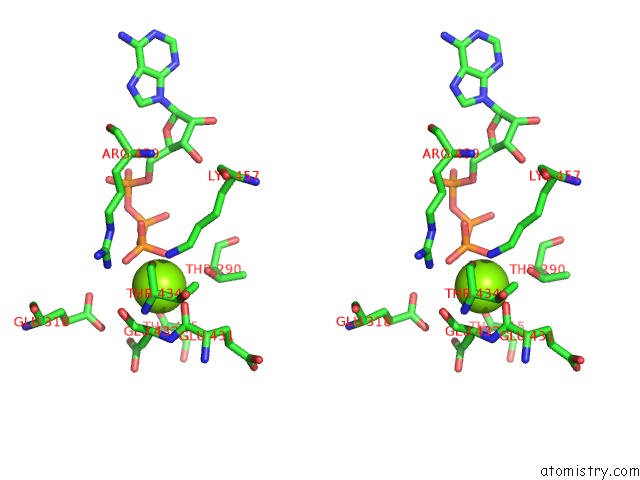
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 4 of Crystal Structure of the Phosphorylation-Site Double Mutant S431E/T432E of the Kaic Circadian Clock Protein within 5.0Å range:
|
Magnesium binding site 5 out of 21 in 3s1a
Go back to
Magnesium binding site 5 out
of 21 in the Crystal Structure of the Phosphorylation-Site Double Mutant S431E/T432E of the Kaic Circadian Clock Protein

Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 5 of Crystal Structure of the Phosphorylation-Site Double Mutant S431E/T432E of the Kaic Circadian Clock Protein within 5.0Å range:
|
Magnesium binding site 6 out of 21 in 3s1a
Go back to
Magnesium binding site 6 out
of 21 in the Crystal Structure of the Phosphorylation-Site Double Mutant S431E/T432E of the Kaic Circadian Clock Protein

Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 6 of Crystal Structure of the Phosphorylation-Site Double Mutant S431E/T432E of the Kaic Circadian Clock Protein within 5.0Å range:
|
Magnesium binding site 7 out of 21 in 3s1a
Go back to
Magnesium binding site 7 out
of 21 in the Crystal Structure of the Phosphorylation-Site Double Mutant S431E/T432E of the Kaic Circadian Clock Protein
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 7 of Crystal Structure of the Phosphorylation-Site Double Mutant S431E/T432E of the Kaic Circadian Clock Protein within 5.0Å range:
|
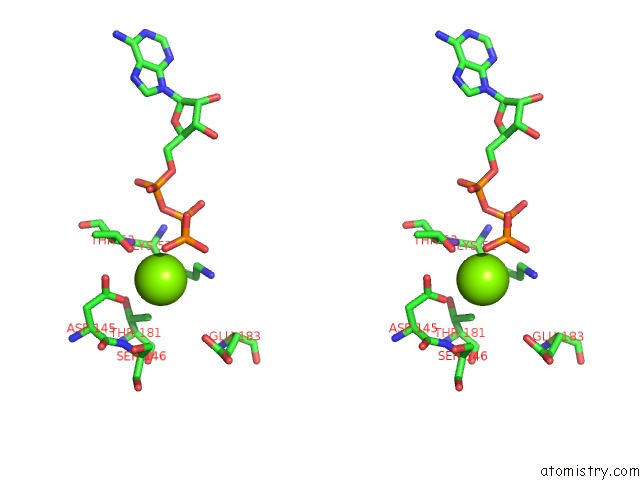
Magnesium binding site 8 out of 21 in 3s1a
Go back to
Magnesium binding site 8 out
of 21 in the Crystal Structure of the Phosphorylation-Site Double Mutant S431E/T432E of the Kaic Circadian Clock Protein

Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 8 of Crystal Structure of the Phosphorylation-Site Double Mutant S431E/T432E of the Kaic Circadian Clock Protein within 5.0Å range:
|
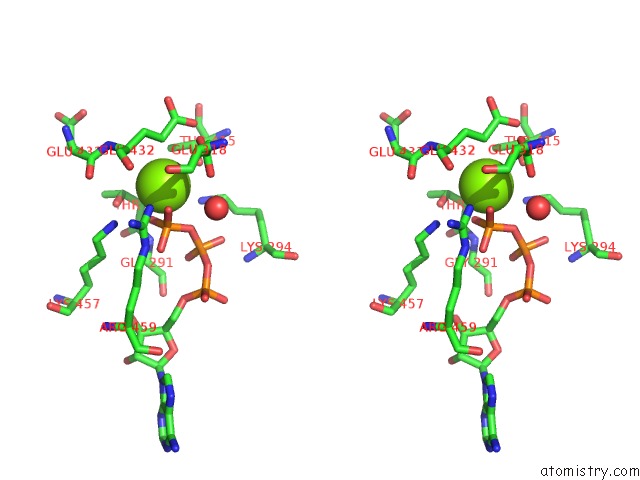
Magnesium binding site 9 out of 21 in 3s1a
Go back to
Magnesium binding site 9 out
of 21 in the Crystal Structure of the Phosphorylation-Site Double Mutant S431E/T432E of the Kaic Circadian Clock Protein

Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 9 of Crystal Structure of the Phosphorylation-Site Double Mutant S431E/T432E of the Kaic Circadian Clock Protein within 5.0Å range:
|
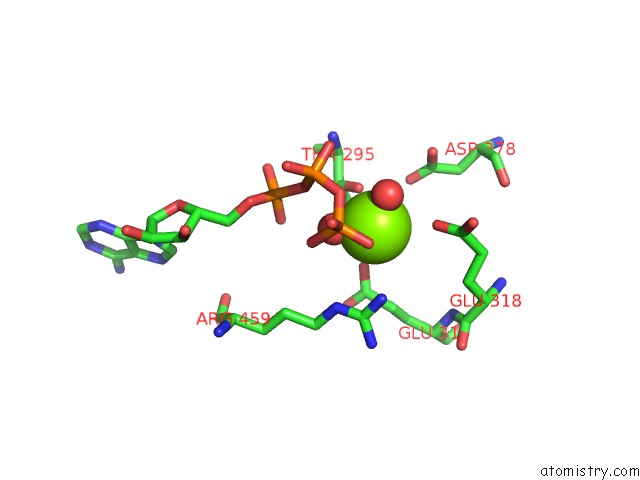
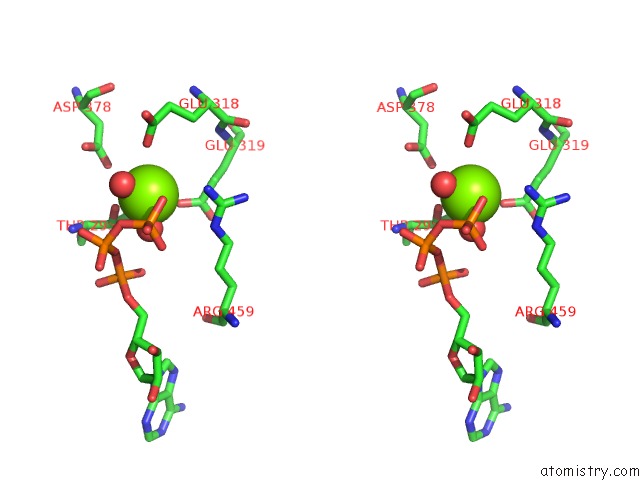
Magnesium binding site 10 out of 21 in 3s1a
Go back to
Magnesium binding site 10 out
of 21 in the Crystal Structure of the Phosphorylation-Site Double Mutant S431E/T432E of the Kaic Circadian Clock Protein
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 10 of Crystal Structure of the Phosphorylation-Site Double Mutant S431E/T432E of the Kaic Circadian Clock Protein within 5.0Å range:
|
Reference:
R.Pattanayek,
D.R.Williams,
G.Rossi,
S.Weigand,
T.Mori,
C.H.Johnson,
P.L.Stewart,
M.Egli.
Combined Saxs/Em Based Models of the S. Elongatus Post-Translational Circadian Oscillator and Its Interactions with the Output His-Kinase Sasa. Plos One V. 6 23697 2011.
ISSN: ESSN 1932-6203
PubMed: 21887298
DOI: 10.1371/JOURNAL.PONE.0023697
Page generated: Thu Aug 15 10:44:06 2024
ISSN: ESSN 1932-6203
PubMed: 21887298
DOI: 10.1371/JOURNAL.PONE.0023697
Last articles
Cl in 5VTKCl in 5VSV
Cl in 5VR0
Cl in 5VTI
Cl in 5VTD
Cl in 5VT7
Cl in 5VSK
Cl in 5VSB
Cl in 5VS7
Cl in 5VPB