Magnesium »
PDB 3rvb-3s8c »
3s3n »
Magnesium in PDB 3s3n: Crystal Structure of the Prototype Foamy Virus (Pfv) S217H Mutant Intasome in Complex with Magnesium and Dolutegravir (S/GSK1349572)
Protein crystallography data
The structure of Crystal Structure of the Prototype Foamy Virus (Pfv) S217H Mutant Intasome in Complex with Magnesium and Dolutegravir (S/GSK1349572), PDB code: 3s3n
was solved by
S.Hare,
P.Cherepanov,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 39.16 / 2.49 |
| Space group | P 41 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 160.050, 160.050, 123.610, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 20.9 / 23.2 |
Other elements in 3s3n:
The structure of Crystal Structure of the Prototype Foamy Virus (Pfv) S217H Mutant Intasome in Complex with Magnesium and Dolutegravir (S/GSK1349572) also contains other interesting chemical elements:
| Fluorine | (F) | 2 atoms |
| Zinc | (Zn) | 1 atom |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Crystal Structure of the Prototype Foamy Virus (Pfv) S217H Mutant Intasome in Complex with Magnesium and Dolutegravir (S/GSK1349572)
(pdb code 3s3n). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 2 binding sites of Magnesium where determined in the Crystal Structure of the Prototype Foamy Virus (Pfv) S217H Mutant Intasome in Complex with Magnesium and Dolutegravir (S/GSK1349572), PDB code: 3s3n:
Jump to Magnesium binding site number: 1; 2;
In total 2 binding sites of Magnesium where determined in the Crystal Structure of the Prototype Foamy Virus (Pfv) S217H Mutant Intasome in Complex with Magnesium and Dolutegravir (S/GSK1349572), PDB code: 3s3n:
Jump to Magnesium binding site number: 1; 2;
Magnesium binding site 1 out of 2 in 3s3n
Go back to
Magnesium binding site 1 out
of 2 in the Crystal Structure of the Prototype Foamy Virus (Pfv) S217H Mutant Intasome in Complex with Magnesium and Dolutegravir (S/GSK1349572)
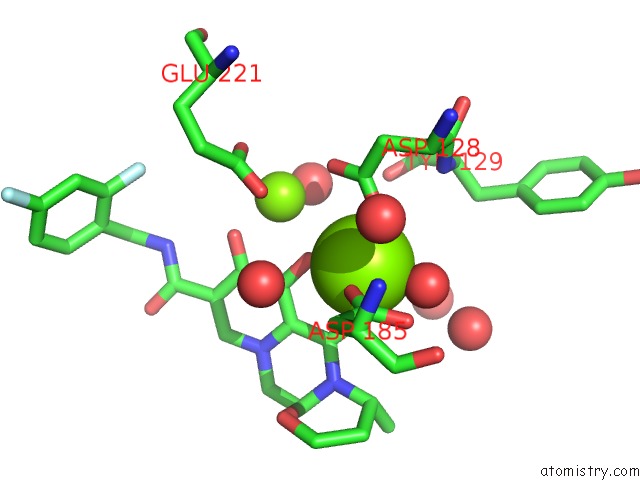
Mono view
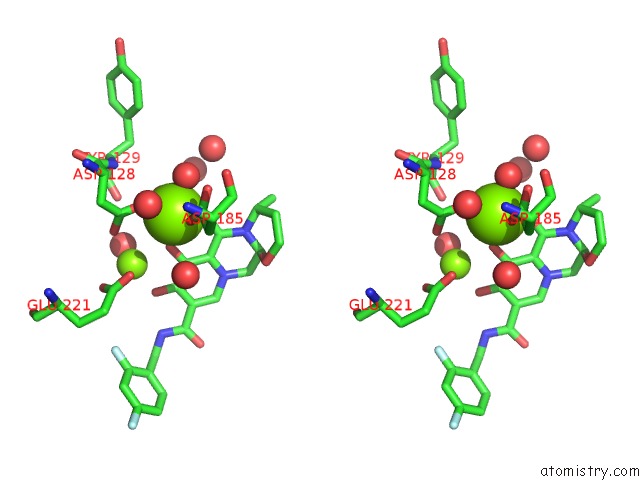
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Crystal Structure of the Prototype Foamy Virus (Pfv) S217H Mutant Intasome in Complex with Magnesium and Dolutegravir (S/GSK1349572) within 5.0Å range:
|
Magnesium binding site 2 out of 2 in 3s3n
Go back to
Magnesium binding site 2 out
of 2 in the Crystal Structure of the Prototype Foamy Virus (Pfv) S217H Mutant Intasome in Complex with Magnesium and Dolutegravir (S/GSK1349572)
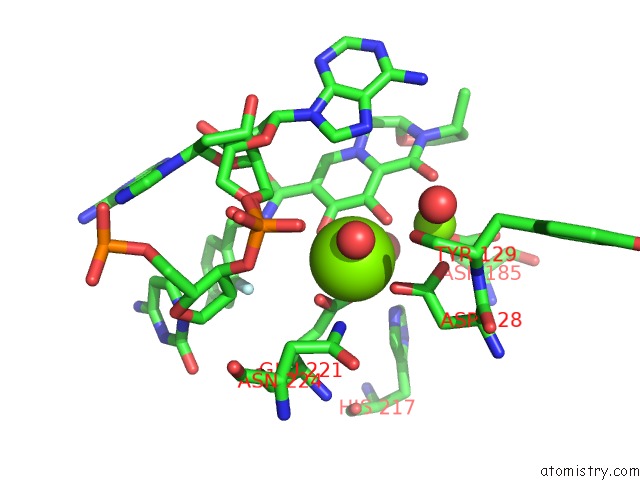
Mono view
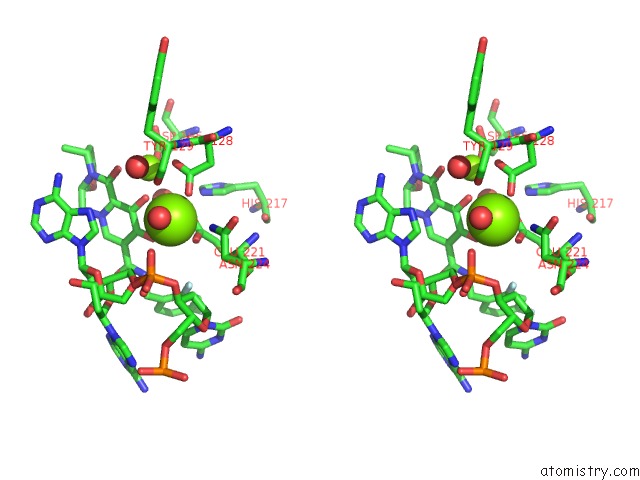
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Crystal Structure of the Prototype Foamy Virus (Pfv) S217H Mutant Intasome in Complex with Magnesium and Dolutegravir (S/GSK1349572) within 5.0Å range:
|
Reference:
S.Hare,
S.J.Smith,
M.Metifiot,
A.Jaxa-Chamiec,
Y.Pommier,
S.H.Hughes,
P.Cherepanov.
Structural and Functional Analyses of the Second-Generation Integrase Strand Transfer Inhibitor Dolutegravir (S/GSK1349572). Mol.Pharmacol. V. 80 565 2011.
ISSN: ISSN 0026-895X
PubMed: 21719464
DOI: 10.1124/MOL.111.073189
Page generated: Thu Aug 15 10:45:19 2024
ISSN: ISSN 0026-895X
PubMed: 21719464
DOI: 10.1124/MOL.111.073189
Last articles
Ca in 5OY0Ca in 5OXG
Ca in 5OXS
Ca in 5OWO
Ca in 5OWR
Ca in 5OWC
Ca in 5OW8
Ca in 5OTJ
Ca in 5OSQ
Ca in 5OTN