Magnesium »
PDB 3umy-3v4i »
3upy »
Magnesium in PDB 3upy: Crystal Structure of the Brucella Abortus Enzyme Catalyzing the First Committed Step of the Methylerythritol 4-Phosphate Pathway.
Protein crystallography data
The structure of Crystal Structure of the Brucella Abortus Enzyme Catalyzing the First Committed Step of the Methylerythritol 4-Phosphate Pathway., PDB code: 3upy
was solved by
B.M.Calisto,
J.Perez-Gil,
I.Fita,
M.Rodriguez-Concepcion,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 39.55 / 1.80 |
| Space group | P 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 49.391, 63.379, 79.334, 68.79, 75.54, 72.07 |
| R / Rfree (%) | 16.3 / 20.4 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Crystal Structure of the Brucella Abortus Enzyme Catalyzing the First Committed Step of the Methylerythritol 4-Phosphate Pathway.
(pdb code 3upy). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 2 binding sites of Magnesium where determined in the Crystal Structure of the Brucella Abortus Enzyme Catalyzing the First Committed Step of the Methylerythritol 4-Phosphate Pathway., PDB code: 3upy:
Jump to Magnesium binding site number: 1; 2;
In total 2 binding sites of Magnesium where determined in the Crystal Structure of the Brucella Abortus Enzyme Catalyzing the First Committed Step of the Methylerythritol 4-Phosphate Pathway., PDB code: 3upy:
Jump to Magnesium binding site number: 1; 2;
Magnesium binding site 1 out of 2 in 3upy
Go back to
Magnesium binding site 1 out
of 2 in the Crystal Structure of the Brucella Abortus Enzyme Catalyzing the First Committed Step of the Methylerythritol 4-Phosphate Pathway.

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Crystal Structure of the Brucella Abortus Enzyme Catalyzing the First Committed Step of the Methylerythritol 4-Phosphate Pathway. within 5.0Å range:
|
Magnesium binding site 2 out of 2 in 3upy
Go back to
Magnesium binding site 2 out
of 2 in the Crystal Structure of the Brucella Abortus Enzyme Catalyzing the First Committed Step of the Methylerythritol 4-Phosphate Pathway.

Mono view
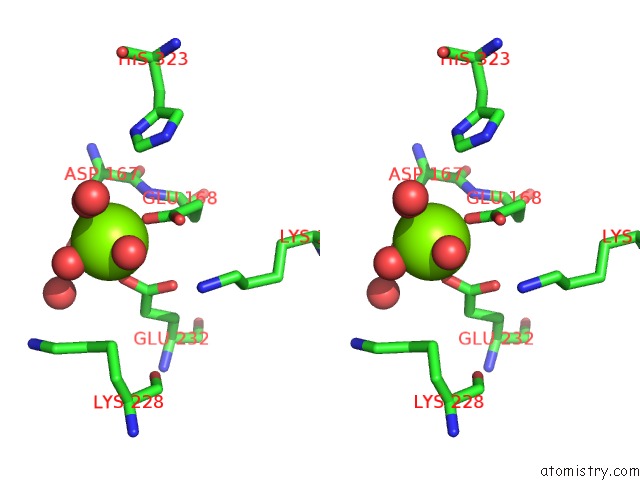
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Crystal Structure of the Brucella Abortus Enzyme Catalyzing the First Committed Step of the Methylerythritol 4-Phosphate Pathway. within 5.0Å range:
|
Reference:
J.Perez-Gil,
B.M.Calisto,
C.Behrendt,
T.Kurz,
I.Fita,
M.Rodriguez-Concepcion.
Crystal Structure of Brucella Abortus Deoxyxylulose-5-Phosphate Reductoisomerase-Like (Drl) Enzyme Involved in Isoprenoid Biosynthesis. J.Biol.Chem. V. 287 15803 2012.
ISSN: ISSN 0021-9258
PubMed: 22442144
DOI: 10.1074/JBC.M112.354811
Page generated: Thu Aug 15 12:37:11 2024
ISSN: ISSN 0021-9258
PubMed: 22442144
DOI: 10.1074/JBC.M112.354811
Last articles
Cl in 7SZECl in 7T0E
Cl in 7SZF
Cl in 7SZQ
Cl in 7SZP
Cl in 7SZH
Cl in 7SZG
Cl in 7SYY
Cl in 7SXL
Cl in 7SZ8