Magnesium »
PDB 3zia-3zx4 »
3zm7 »
Magnesium in PDB 3zm7: Crystal Structure of the Atpase Region of Mycobacterium Tuberculosis Gyrb with Amppcp
Enzymatic activity of Crystal Structure of the Atpase Region of Mycobacterium Tuberculosis Gyrb with Amppcp
All present enzymatic activity of Crystal Structure of the Atpase Region of Mycobacterium Tuberculosis Gyrb with Amppcp:
5.99.1.3;
5.99.1.3;
Protein crystallography data
The structure of Crystal Structure of the Atpase Region of Mycobacterium Tuberculosis Gyrb with Amppcp, PDB code: 3zm7
was solved by
A.Agrawal,
M.Roue,
C.Spitzfaden,
S.Petrella,
A.Aubry,
C.Volker,
D.Mossakowska,
M.Hann,
B.Bax,
C.Mayer,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 25.00 / 3.30 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 78.615, 171.916, 109.230, 90.00, 110.22, 90.00 |
| R / Rfree (%) | 19.57 / 22.31 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Crystal Structure of the Atpase Region of Mycobacterium Tuberculosis Gyrb with Amppcp
(pdb code 3zm7). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 6 binding sites of Magnesium where determined in the Crystal Structure of the Atpase Region of Mycobacterium Tuberculosis Gyrb with Amppcp, PDB code: 3zm7:
Jump to Magnesium binding site number: 1; 2; 3; 4; 5; 6;
In total 6 binding sites of Magnesium where determined in the Crystal Structure of the Atpase Region of Mycobacterium Tuberculosis Gyrb with Amppcp, PDB code: 3zm7:
Jump to Magnesium binding site number: 1; 2; 3; 4; 5; 6;
Magnesium binding site 1 out of 6 in 3zm7
Go back to
Magnesium binding site 1 out
of 6 in the Crystal Structure of the Atpase Region of Mycobacterium Tuberculosis Gyrb with Amppcp
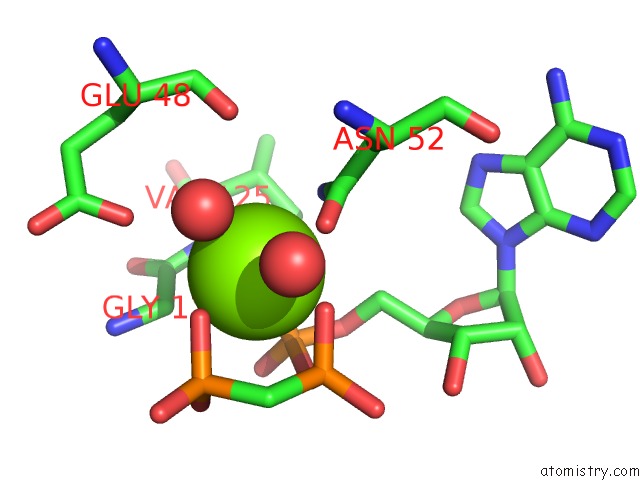
Mono view
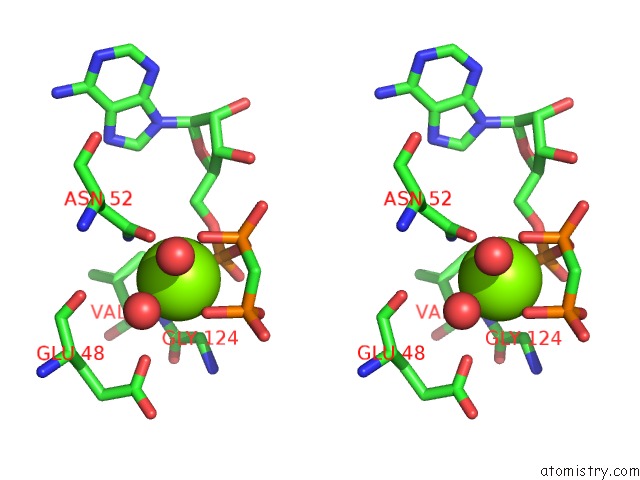
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Crystal Structure of the Atpase Region of Mycobacterium Tuberculosis Gyrb with Amppcp within 5.0Å range:
|
Magnesium binding site 2 out of 6 in 3zm7
Go back to
Magnesium binding site 2 out
of 6 in the Crystal Structure of the Atpase Region of Mycobacterium Tuberculosis Gyrb with Amppcp
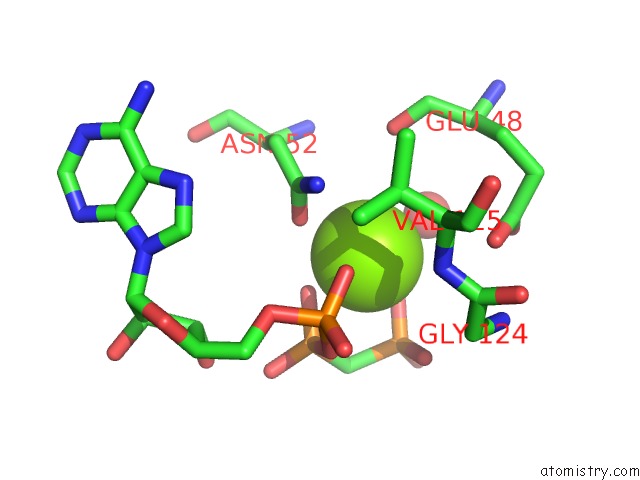
Mono view
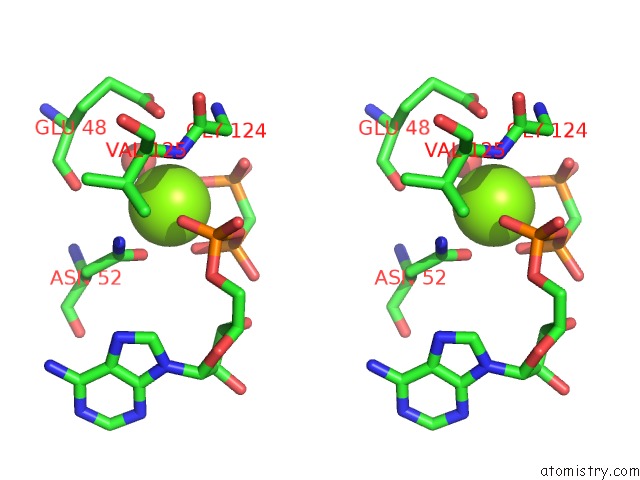
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Crystal Structure of the Atpase Region of Mycobacterium Tuberculosis Gyrb with Amppcp within 5.0Å range:
|
Magnesium binding site 3 out of 6 in 3zm7
Go back to
Magnesium binding site 3 out
of 6 in the Crystal Structure of the Atpase Region of Mycobacterium Tuberculosis Gyrb with Amppcp
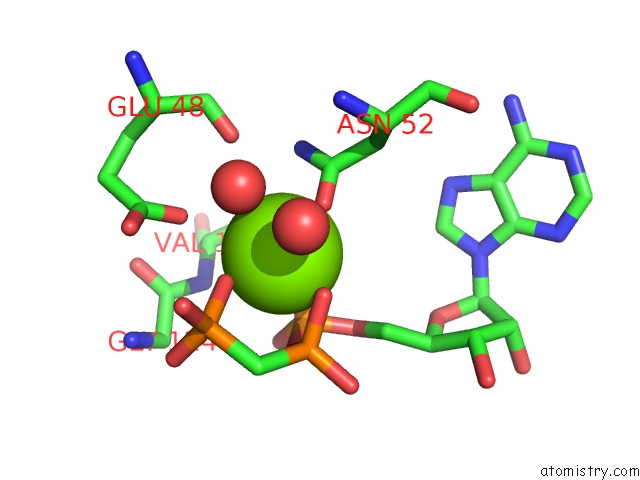
Mono view
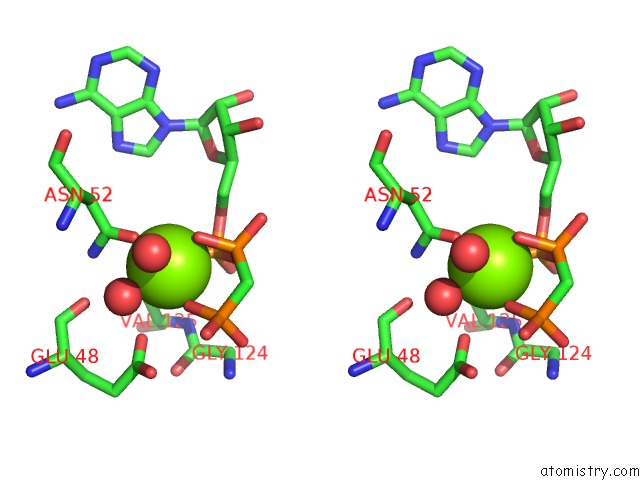
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 3 of Crystal Structure of the Atpase Region of Mycobacterium Tuberculosis Gyrb with Amppcp within 5.0Å range:
|
Magnesium binding site 4 out of 6 in 3zm7
Go back to
Magnesium binding site 4 out
of 6 in the Crystal Structure of the Atpase Region of Mycobacterium Tuberculosis Gyrb with Amppcp
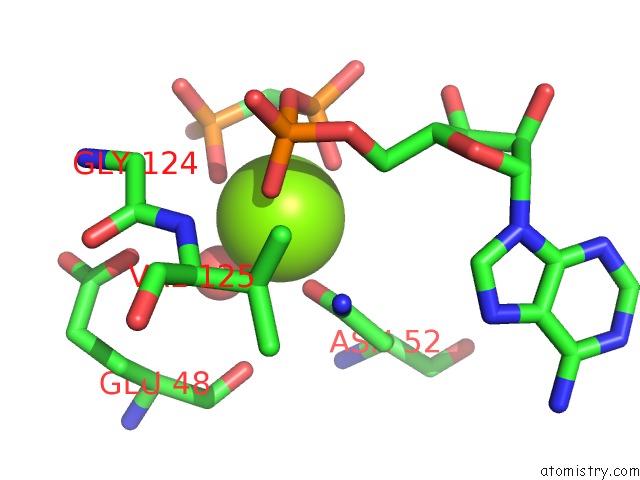
Mono view
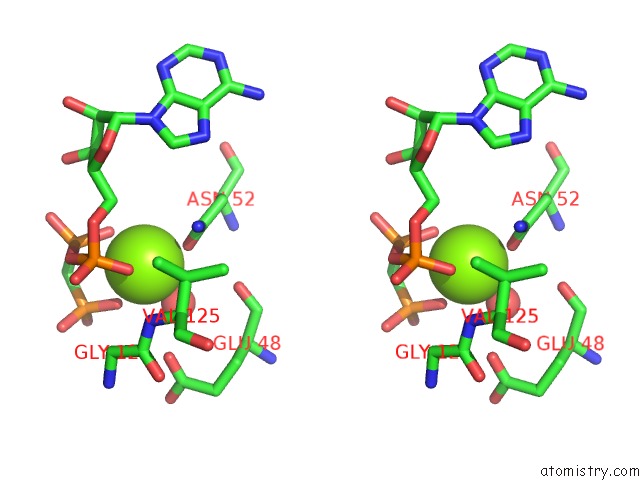
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 4 of Crystal Structure of the Atpase Region of Mycobacterium Tuberculosis Gyrb with Amppcp within 5.0Å range:
|
Magnesium binding site 5 out of 6 in 3zm7
Go back to
Magnesium binding site 5 out
of 6 in the Crystal Structure of the Atpase Region of Mycobacterium Tuberculosis Gyrb with Amppcp
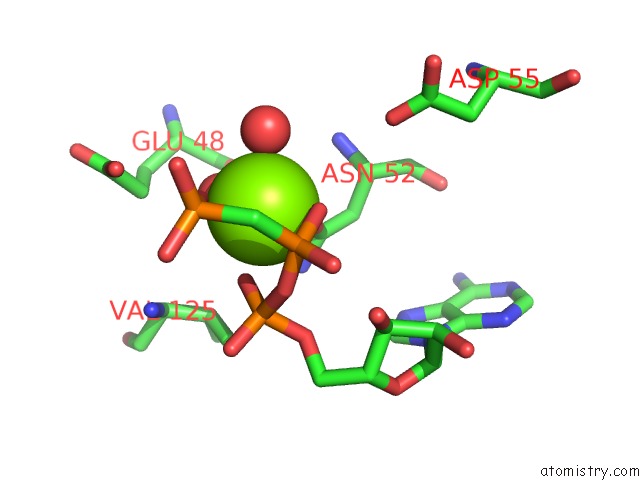
Mono view
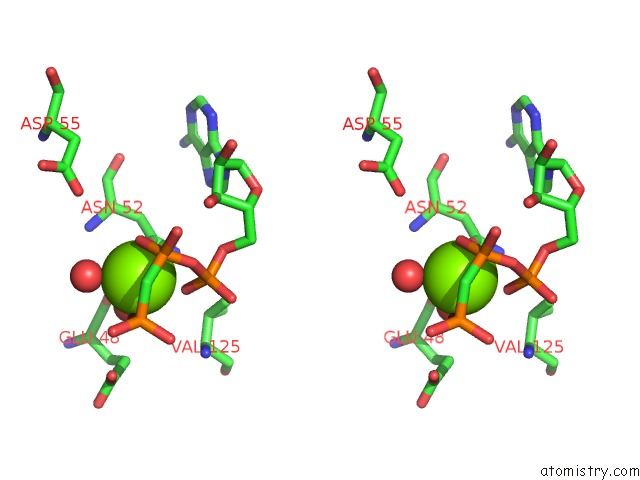
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 5 of Crystal Structure of the Atpase Region of Mycobacterium Tuberculosis Gyrb with Amppcp within 5.0Å range:
|
Magnesium binding site 6 out of 6 in 3zm7
Go back to
Magnesium binding site 6 out
of 6 in the Crystal Structure of the Atpase Region of Mycobacterium Tuberculosis Gyrb with Amppcp
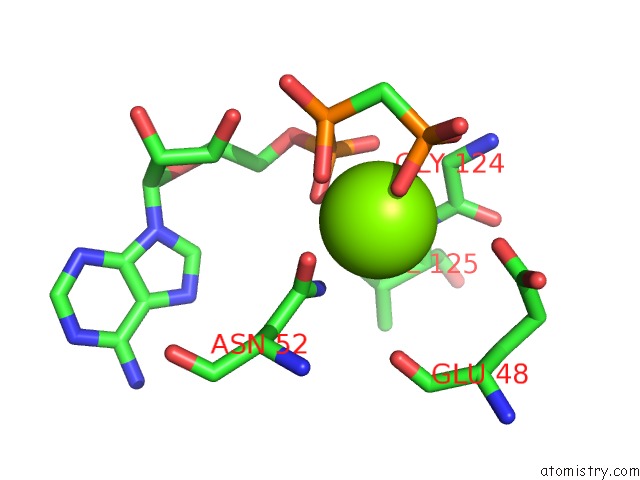
Mono view
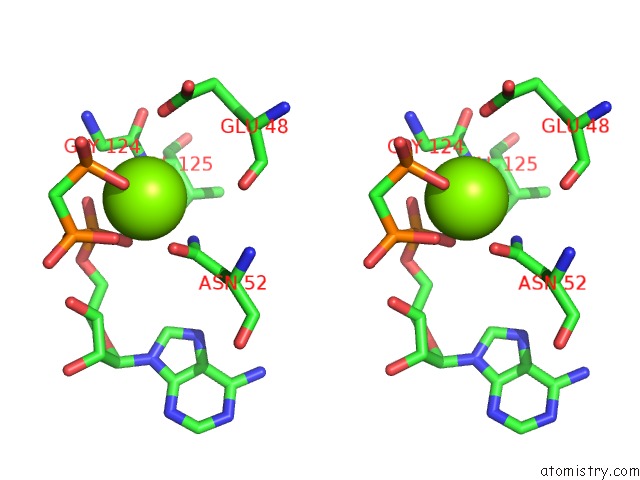
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 6 of Crystal Structure of the Atpase Region of Mycobacterium Tuberculosis Gyrb with Amppcp within 5.0Å range:
|
Reference:
A.Agrawal,
M.Roue,
C.Spitzfaden,
S.Petrella,
A.Aubry,
M.M.Hann,
B.Bax,
C.Mayer.
Mycobacterium Tuberculosis Dna Gyrase Atpase Domain Structures Suggest A Dissociative Mechanism That Explains How Atp Hydrolysis Is Coupled to Domain Motion. Biochem.J. V. 456 263 2013.
ISSN: ISSN 0264-6021
PubMed: 24015710
DOI: 10.1042/BJ20130538
Page generated: Mon Aug 11 05:17:00 2025
ISSN: ISSN 0264-6021
PubMed: 24015710
DOI: 10.1042/BJ20130538
Last articles
Mg in 4OMFMg in 4OO1
Mg in 4OKM
Mg in 4OLS
Mg in 4OL0
Mg in 4OKK
Mg in 4OKJ
Mg in 4OKQ
Mg in 4OK9
Mg in 4OKE