Magnesium »
PDB 4brs-4c2z »
4bwa »
Magnesium in PDB 4bwa: Pylrs Y306G, Y384F, I405R Mutant in Complex with Adenylated Norbornene
Enzymatic activity of Pylrs Y306G, Y384F, I405R Mutant in Complex with Adenylated Norbornene
All present enzymatic activity of Pylrs Y306G, Y384F, I405R Mutant in Complex with Adenylated Norbornene:
6.1.1.26;
6.1.1.26;
Protein crystallography data
The structure of Pylrs Y306G, Y384F, I405R Mutant in Complex with Adenylated Norbornene, PDB code: 4bwa
was solved by
S.Schneider,
M.Vrabel,
M.J.Gattner,
V.Fluegel,
V.Lopez-Carillo,
T.Carell,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 45.42 / 2.45 |
| Space group | P 64 |
| Cell size a, b, c (Å), α, β, γ (°) | 104.800, 104.800, 72.860, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 18.281 / 21.369 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Pylrs Y306G, Y384F, I405R Mutant in Complex with Adenylated Norbornene
(pdb code 4bwa). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total only one binding site of Magnesium was determined in the Pylrs Y306G, Y384F, I405R Mutant in Complex with Adenylated Norbornene, PDB code: 4bwa:
In total only one binding site of Magnesium was determined in the Pylrs Y306G, Y384F, I405R Mutant in Complex with Adenylated Norbornene, PDB code: 4bwa:
Magnesium binding site 1 out of 1 in 4bwa
Go back to
Magnesium binding site 1 out
of 1 in the Pylrs Y306G, Y384F, I405R Mutant in Complex with Adenylated Norbornene
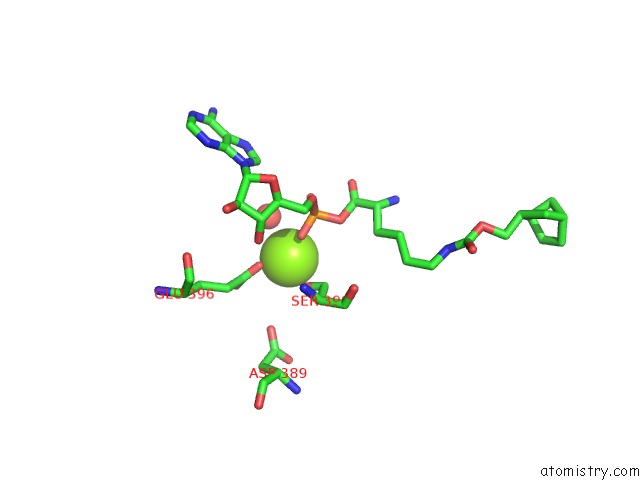
Mono view
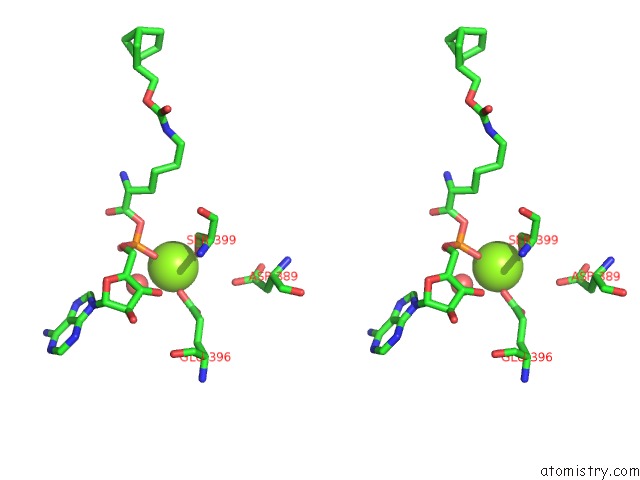
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Pylrs Y306G, Y384F, I405R Mutant in Complex with Adenylated Norbornene within 5.0Å range:
|
Reference:
S.Schneider,
M.J.Gattner,
M.Vrabel,
V.Flugel,
V.Lopez-Carrillo,
S.Prill,
T.Carell.
Structural Insights Into Incorporation of Norbornene Amino Acids For Click Modification of Proteins Chem.Bio.Chem. V. 14 2114 2013.
ISSN: ISSN 1439-4227
PubMed: 24027216
DOI: 10.1002/CBIC.201300435
Page generated: Mon Aug 11 07:06:22 2025
ISSN: ISSN 1439-4227
PubMed: 24027216
DOI: 10.1002/CBIC.201300435
Last articles
Mg in 5B48Mg in 5B47
Mg in 5B46
Mg in 5B30
Mg in 5B2Z
Mg in 5B2T
Mg in 5B2S
Mg in 5B2R
Mg in 5B25
Mg in 5AYR