Magnesium »
PDB 4d5e-4dhf »
4dat »
Magnesium in PDB 4dat: Structure of 14-3-3 Sigma in Complex with PADI6 14-3-3 Binding Motif II
Enzymatic activity of Structure of 14-3-3 Sigma in Complex with PADI6 14-3-3 Binding Motif II
All present enzymatic activity of Structure of 14-3-3 Sigma in Complex with PADI6 14-3-3 Binding Motif II:
3.5.3.15;
3.5.3.15;
Protein crystallography data
The structure of Structure of 14-3-3 Sigma in Complex with PADI6 14-3-3 Binding Motif II, PDB code: 4dat
was solved by
R.Rose,
M.Rose,
C.Ottmann,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 45.58 / 1.40 |
| Space group | C 2 2 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 82.220, 112.360, 62.730, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 14.7 / 18.3 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Structure of 14-3-3 Sigma in Complex with PADI6 14-3-3 Binding Motif II
(pdb code 4dat). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total only one binding site of Magnesium was determined in the Structure of 14-3-3 Sigma in Complex with PADI6 14-3-3 Binding Motif II, PDB code: 4dat:
In total only one binding site of Magnesium was determined in the Structure of 14-3-3 Sigma in Complex with PADI6 14-3-3 Binding Motif II, PDB code: 4dat:
Magnesium binding site 1 out of 1 in 4dat
Go back to
Magnesium binding site 1 out
of 1 in the Structure of 14-3-3 Sigma in Complex with PADI6 14-3-3 Binding Motif II
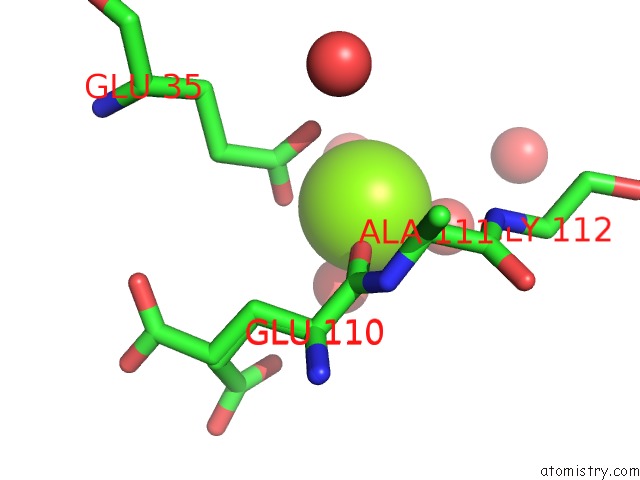
Mono view
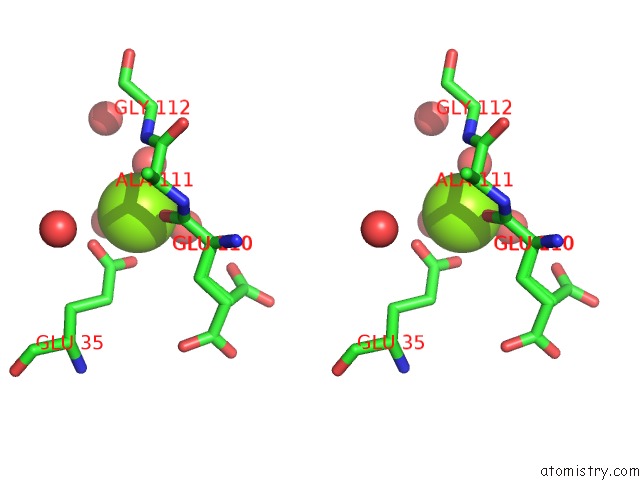
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Structure of 14-3-3 Sigma in Complex with PADI6 14-3-3 Binding Motif II within 5.0Å range:
|
Reference:
R.Rose,
M.Rose,
C.Ottmann.
Identification and Structural Characterization of Two 14-3-3 Binding Sites in the Human Peptidylarginine Deiminase Type VI. J.Struct.Biol. V. 180 65 2012.
ISSN: ISSN 1047-8477
PubMed: 22634725
DOI: 10.1016/J.JSB.2012.05.010
Page generated: Mon Aug 11 07:26:01 2025
ISSN: ISSN 1047-8477
PubMed: 22634725
DOI: 10.1016/J.JSB.2012.05.010
Last articles
Mg in 4JI6Mg in 4JJS
Mg in 4JJ2
Mg in 4JIW
Mg in 4JIV
Mg in 4JIB
Mg in 4JI4
Mg in 4JI5
Mg in 4JI1
Mg in 4JI0