Magnesium »
PDB 4gyz-4h9l »
4h2h »
Magnesium in PDB 4h2h: Crystal Structure of An Enolase (Mandalate Racemase Subgroup, Target Efi-502101) From Pelagibaca Bermudensis HTCC2601, with Bound Mg and L-4-Hydroxyproline Betaine (Betonicine)
Protein crystallography data
The structure of Crystal Structure of An Enolase (Mandalate Racemase Subgroup, Target Efi-502101) From Pelagibaca Bermudensis HTCC2601, with Bound Mg and L-4-Hydroxyproline Betaine (Betonicine), PDB code: 4h2h
was solved by
M.W.Vetting,
L.L.Morisco,
S.R.Wasserman,
S.Sojitra,
H.J.Imker,
J.A.Gerlt,
S.C.Almo,
Enzyme Function Initiative (Efi),
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 25.68 / 1.70 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 91.035, 152.824, 113.027, 90.00, 105.20, 90.00 |
| R / Rfree (%) | 15.9 / 19.2 |
Other elements in 4h2h:
The structure of Crystal Structure of An Enolase (Mandalate Racemase Subgroup, Target Efi-502101) From Pelagibaca Bermudensis HTCC2601, with Bound Mg and L-4-Hydroxyproline Betaine (Betonicine) also contains other interesting chemical elements:
| Nickel | (Ni) | 2 atoms |
| Iodine | (I) | 61 atoms |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Crystal Structure of An Enolase (Mandalate Racemase Subgroup, Target Efi-502101) From Pelagibaca Bermudensis HTCC2601, with Bound Mg and L-4-Hydroxyproline Betaine (Betonicine)
(pdb code 4h2h). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 8 binding sites of Magnesium where determined in the Crystal Structure of An Enolase (Mandalate Racemase Subgroup, Target Efi-502101) From Pelagibaca Bermudensis HTCC2601, with Bound Mg and L-4-Hydroxyproline Betaine (Betonicine), PDB code: 4h2h:
Jump to Magnesium binding site number: 1; 2; 3; 4; 5; 6; 7; 8;
In total 8 binding sites of Magnesium where determined in the Crystal Structure of An Enolase (Mandalate Racemase Subgroup, Target Efi-502101) From Pelagibaca Bermudensis HTCC2601, with Bound Mg and L-4-Hydroxyproline Betaine (Betonicine), PDB code: 4h2h:
Jump to Magnesium binding site number: 1; 2; 3; 4; 5; 6; 7; 8;
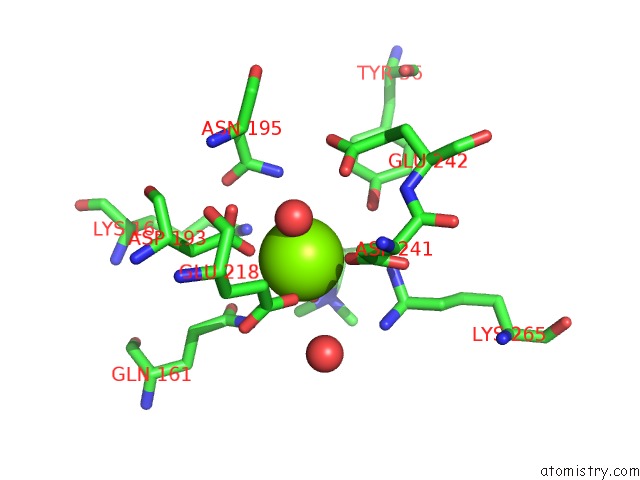
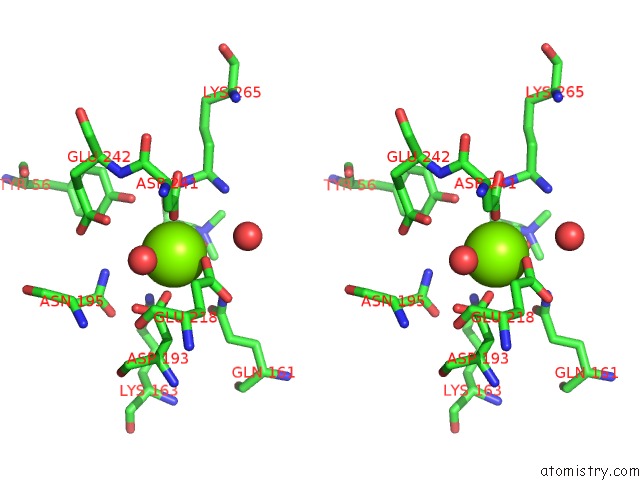
Magnesium binding site 1 out of 8 in 4h2h
Go back to
Magnesium binding site 1 out
of 8 in the Crystal Structure of An Enolase (Mandalate Racemase Subgroup, Target Efi-502101) From Pelagibaca Bermudensis HTCC2601, with Bound Mg and L-4-Hydroxyproline Betaine (Betonicine)
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Crystal Structure of An Enolase (Mandalate Racemase Subgroup, Target Efi-502101) From Pelagibaca Bermudensis HTCC2601, with Bound Mg and L-4-Hydroxyproline Betaine (Betonicine) within 5.0Å range:
|
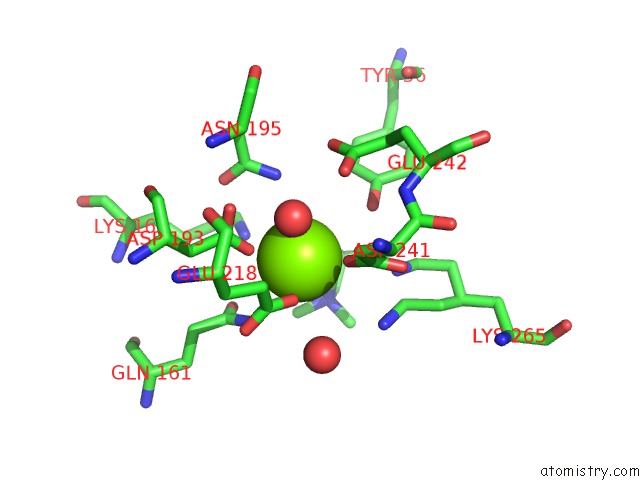
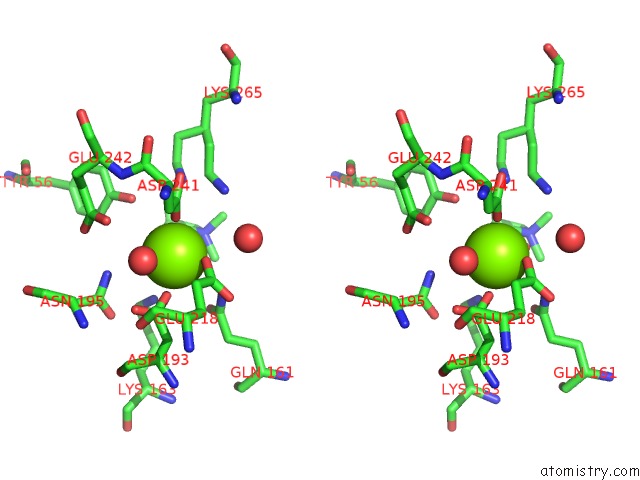
Magnesium binding site 2 out of 8 in 4h2h
Go back to
Magnesium binding site 2 out
of 8 in the Crystal Structure of An Enolase (Mandalate Racemase Subgroup, Target Efi-502101) From Pelagibaca Bermudensis HTCC2601, with Bound Mg and L-4-Hydroxyproline Betaine (Betonicine)
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Crystal Structure of An Enolase (Mandalate Racemase Subgroup, Target Efi-502101) From Pelagibaca Bermudensis HTCC2601, with Bound Mg and L-4-Hydroxyproline Betaine (Betonicine) within 5.0Å range:
|
Magnesium binding site 3 out of 8 in 4h2h
Go back to
Magnesium binding site 3 out
of 8 in the Crystal Structure of An Enolase (Mandalate Racemase Subgroup, Target Efi-502101) From Pelagibaca Bermudensis HTCC2601, with Bound Mg and L-4-Hydroxyproline Betaine (Betonicine)
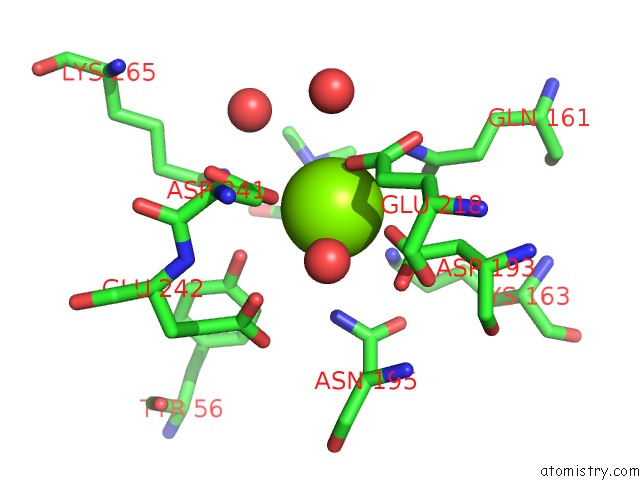
Mono view
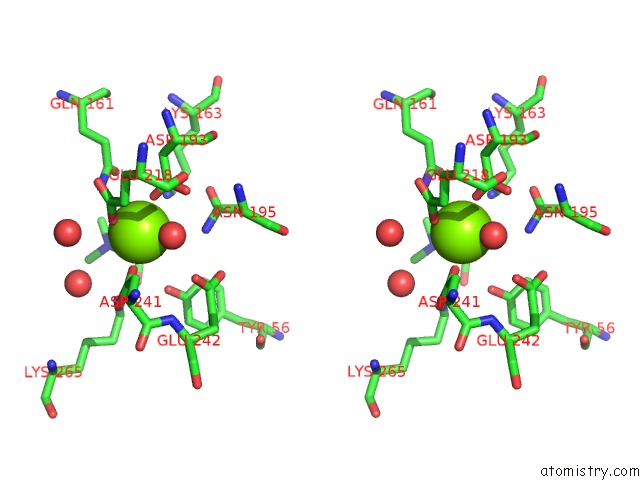
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 3 of Crystal Structure of An Enolase (Mandalate Racemase Subgroup, Target Efi-502101) From Pelagibaca Bermudensis HTCC2601, with Bound Mg and L-4-Hydroxyproline Betaine (Betonicine) within 5.0Å range:
|
Magnesium binding site 4 out of 8 in 4h2h
Go back to
Magnesium binding site 4 out
of 8 in the Crystal Structure of An Enolase (Mandalate Racemase Subgroup, Target Efi-502101) From Pelagibaca Bermudensis HTCC2601, with Bound Mg and L-4-Hydroxyproline Betaine (Betonicine)
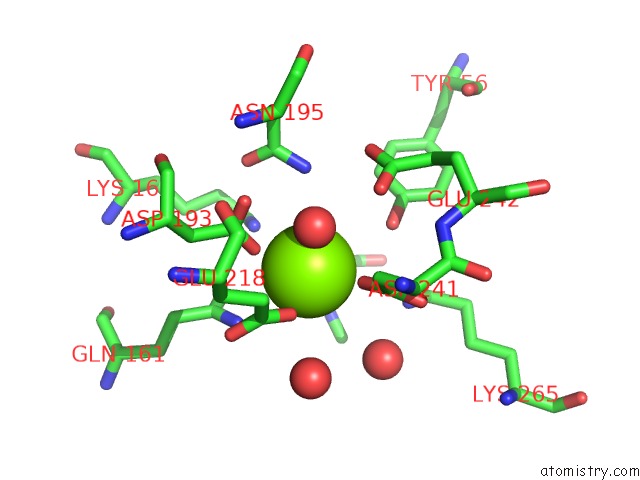
Mono view
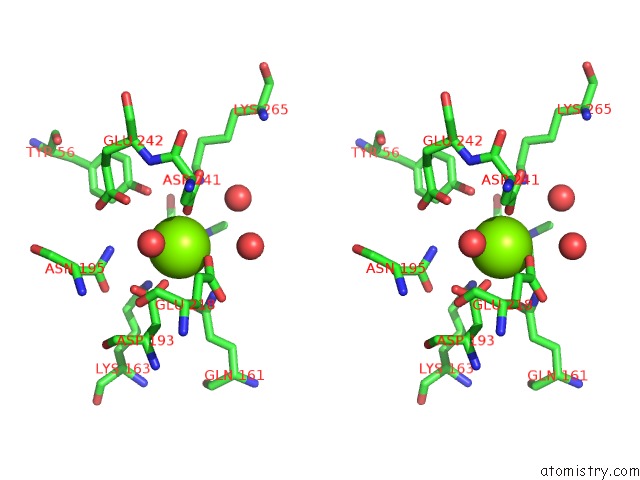
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 4 of Crystal Structure of An Enolase (Mandalate Racemase Subgroup, Target Efi-502101) From Pelagibaca Bermudensis HTCC2601, with Bound Mg and L-4-Hydroxyproline Betaine (Betonicine) within 5.0Å range:
|
Magnesium binding site 5 out of 8 in 4h2h
Go back to
Magnesium binding site 5 out
of 8 in the Crystal Structure of An Enolase (Mandalate Racemase Subgroup, Target Efi-502101) From Pelagibaca Bermudensis HTCC2601, with Bound Mg and L-4-Hydroxyproline Betaine (Betonicine)
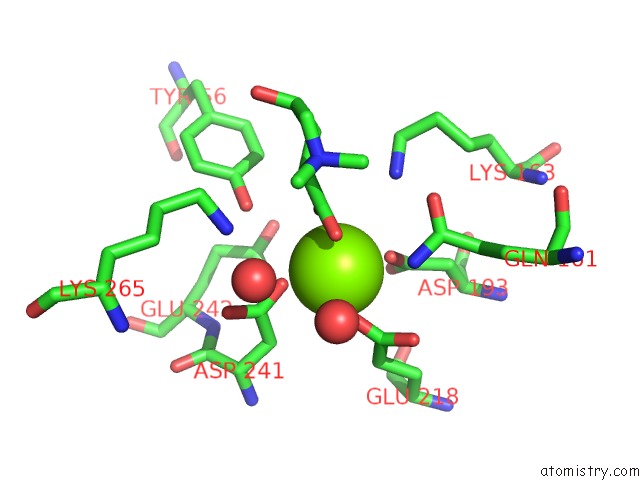
Mono view
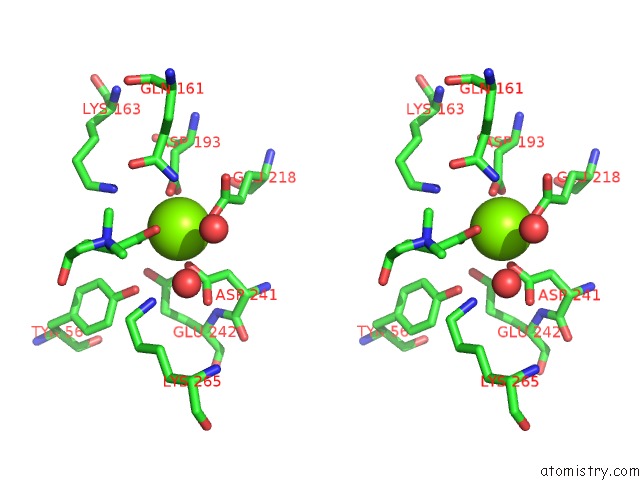
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 5 of Crystal Structure of An Enolase (Mandalate Racemase Subgroup, Target Efi-502101) From Pelagibaca Bermudensis HTCC2601, with Bound Mg and L-4-Hydroxyproline Betaine (Betonicine) within 5.0Å range:
|
Magnesium binding site 6 out of 8 in 4h2h
Go back to
Magnesium binding site 6 out
of 8 in the Crystal Structure of An Enolase (Mandalate Racemase Subgroup, Target Efi-502101) From Pelagibaca Bermudensis HTCC2601, with Bound Mg and L-4-Hydroxyproline Betaine (Betonicine)
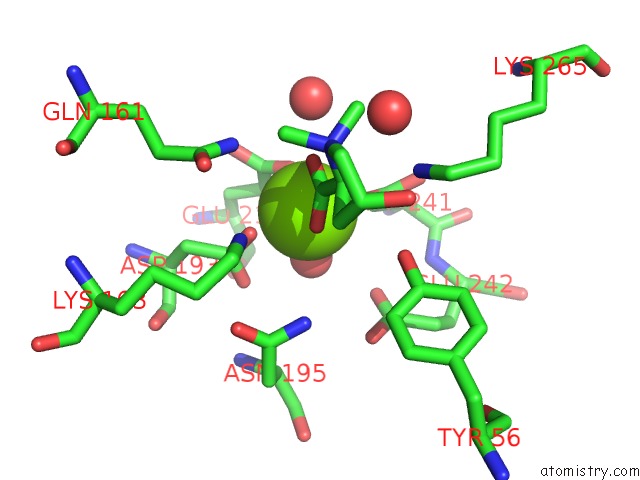
Mono view
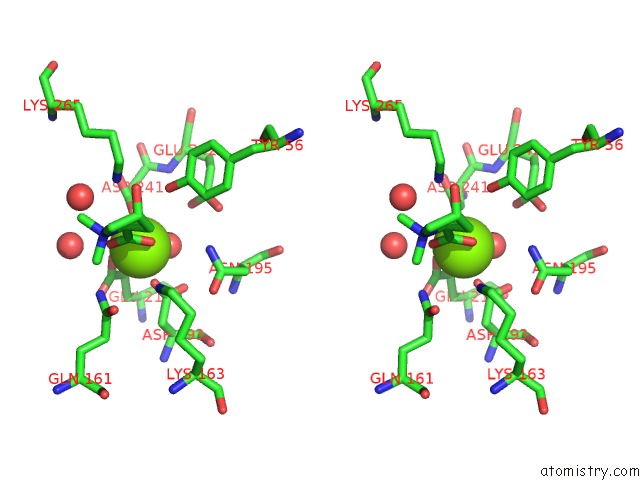
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 6 of Crystal Structure of An Enolase (Mandalate Racemase Subgroup, Target Efi-502101) From Pelagibaca Bermudensis HTCC2601, with Bound Mg and L-4-Hydroxyproline Betaine (Betonicine) within 5.0Å range:
|
Magnesium binding site 7 out of 8 in 4h2h
Go back to
Magnesium binding site 7 out
of 8 in the Crystal Structure of An Enolase (Mandalate Racemase Subgroup, Target Efi-502101) From Pelagibaca Bermudensis HTCC2601, with Bound Mg and L-4-Hydroxyproline Betaine (Betonicine)
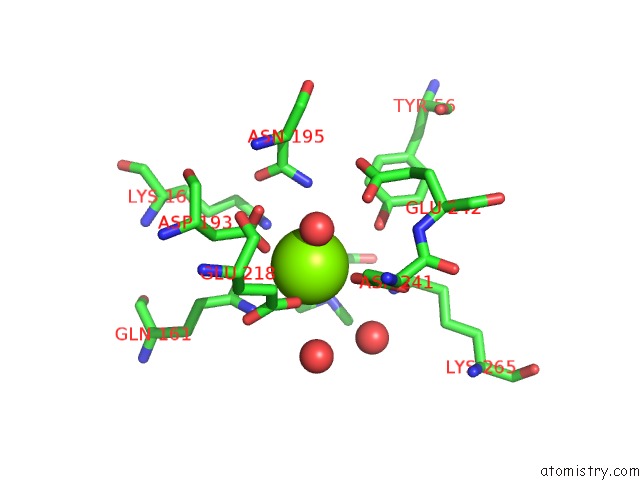
Mono view
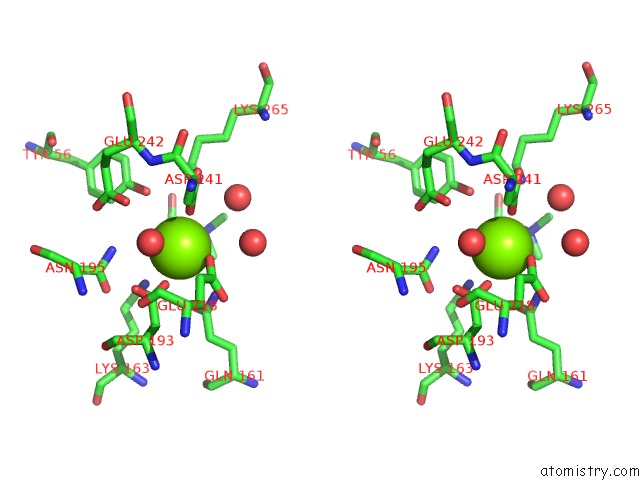
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 7 of Crystal Structure of An Enolase (Mandalate Racemase Subgroup, Target Efi-502101) From Pelagibaca Bermudensis HTCC2601, with Bound Mg and L-4-Hydroxyproline Betaine (Betonicine) within 5.0Å range:
|
Magnesium binding site 8 out of 8 in 4h2h
Go back to
Magnesium binding site 8 out
of 8 in the Crystal Structure of An Enolase (Mandalate Racemase Subgroup, Target Efi-502101) From Pelagibaca Bermudensis HTCC2601, with Bound Mg and L-4-Hydroxyproline Betaine (Betonicine)
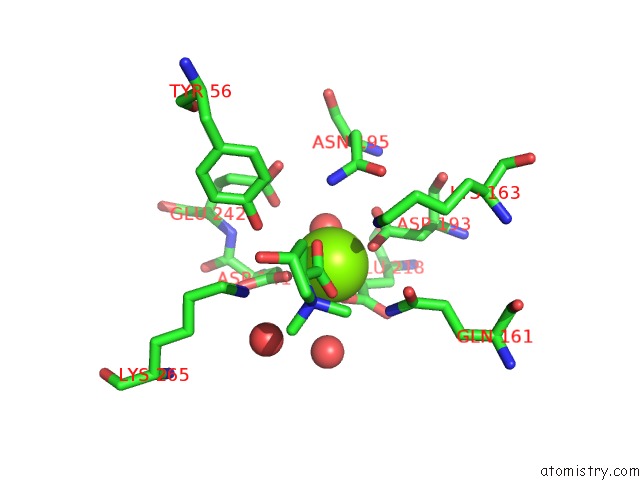
Mono view
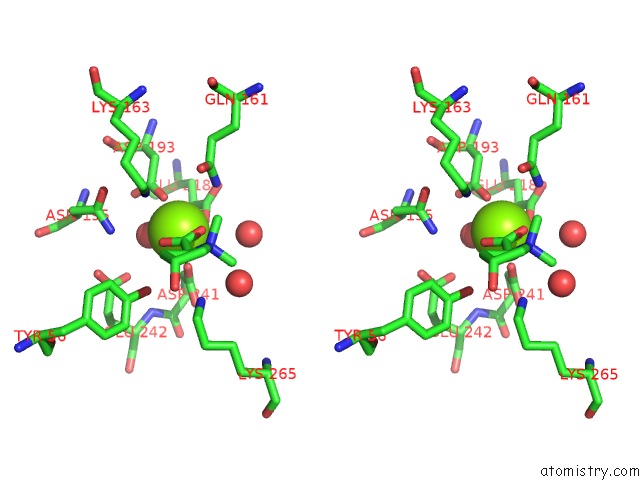
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 8 of Crystal Structure of An Enolase (Mandalate Racemase Subgroup, Target Efi-502101) From Pelagibaca Bermudensis HTCC2601, with Bound Mg and L-4-Hydroxyproline Betaine (Betonicine) within 5.0Å range:
|
Reference:
S.Zhao,
R.Kumar,
A.Sakai,
M.W.Vetting,
B.M.Wood,
S.Brown,
J.B.Bonanno,
B.S.Hillerich,
R.D.Seidel,
P.C.Babbitt,
S.C.Almo,
J.V.Sweedler,
J.A.Gerlt,
J.E.Cronan,
M.P.Jacobson.
Discovery of New Enzymes and Metabolic Pathways By Using Structure and Genome Context. Nature V. 502 698 2013.
ISSN: ISSN 0028-0836
PubMed: 24056934
DOI: 10.1038/NATURE12576
Page generated: Fri Aug 16 16:08:40 2024
ISSN: ISSN 0028-0836
PubMed: 24056934
DOI: 10.1038/NATURE12576
Last articles
F in 4GHIF in 4GG7
F in 4GG5
F in 4GE6
F in 4GE5
F in 4GCA
F in 4GE2
F in 4G9C
F in 4GAB
F in 4GA8