Magnesium »
PDB 4ibd-4iir »
4ifw »
Magnesium in PDB 4ifw: Crystal Structure of Treponema Pallidum TP0796 Flavin Trafficking Protein, Adp Inhibited Form
Protein crystallography data
The structure of Crystal Structure of Treponema Pallidum TP0796 Flavin Trafficking Protein, Adp Inhibited Form, PDB code: 4ifw
was solved by
D.R.Tomchick,
C.A.Brautigam,
R.K.Deka,
M.V.Norgard,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 29.65 / 2.30 |
| Space group | C 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 116.829, 47.252, 57.643, 90.00, 102.03, 90.00 |
| R / Rfree (%) | 17.7 / 23.8 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Crystal Structure of Treponema Pallidum TP0796 Flavin Trafficking Protein, Adp Inhibited Form
(pdb code 4ifw). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 2 binding sites of Magnesium where determined in the Crystal Structure of Treponema Pallidum TP0796 Flavin Trafficking Protein, Adp Inhibited Form, PDB code: 4ifw:
Jump to Magnesium binding site number: 1; 2;
In total 2 binding sites of Magnesium where determined in the Crystal Structure of Treponema Pallidum TP0796 Flavin Trafficking Protein, Adp Inhibited Form, PDB code: 4ifw:
Jump to Magnesium binding site number: 1; 2;
Magnesium binding site 1 out of 2 in 4ifw
Go back to
Magnesium binding site 1 out
of 2 in the Crystal Structure of Treponema Pallidum TP0796 Flavin Trafficking Protein, Adp Inhibited Form
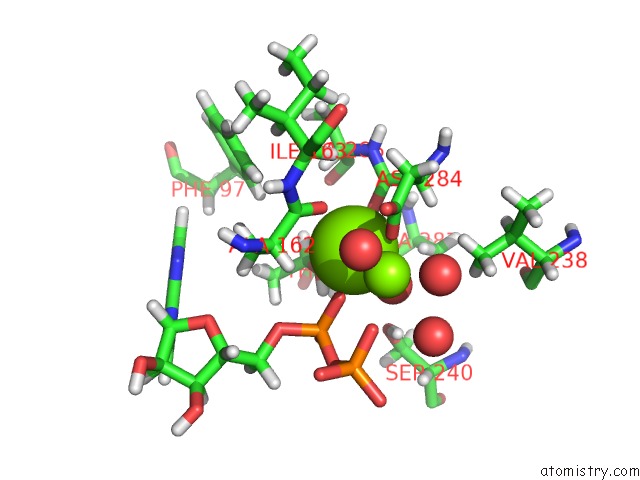
Mono view
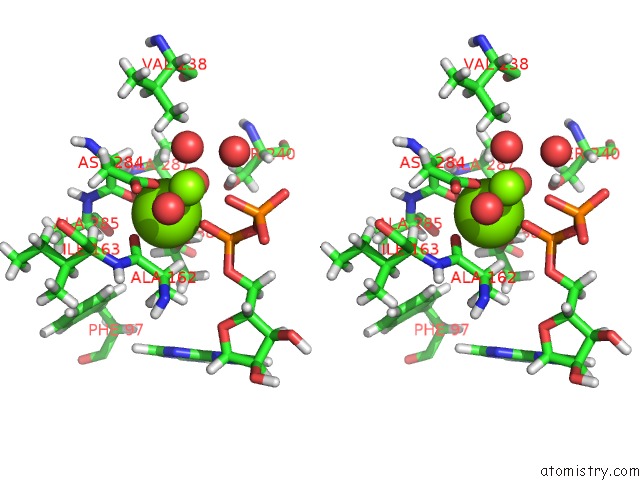
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Crystal Structure of Treponema Pallidum TP0796 Flavin Trafficking Protein, Adp Inhibited Form within 5.0Å range:
|
Magnesium binding site 2 out of 2 in 4ifw
Go back to
Magnesium binding site 2 out
of 2 in the Crystal Structure of Treponema Pallidum TP0796 Flavin Trafficking Protein, Adp Inhibited Form
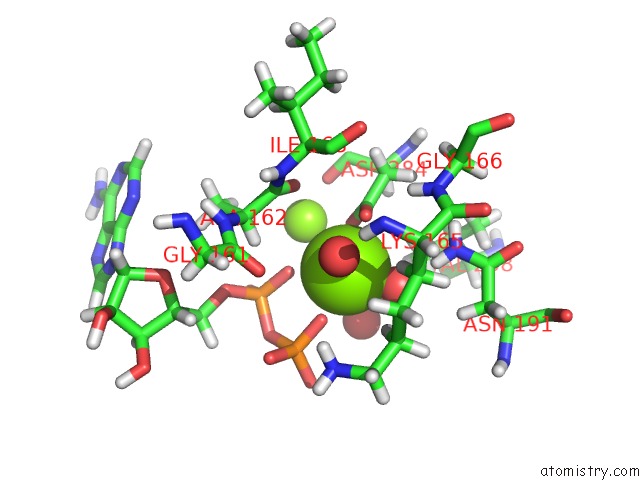
Mono view
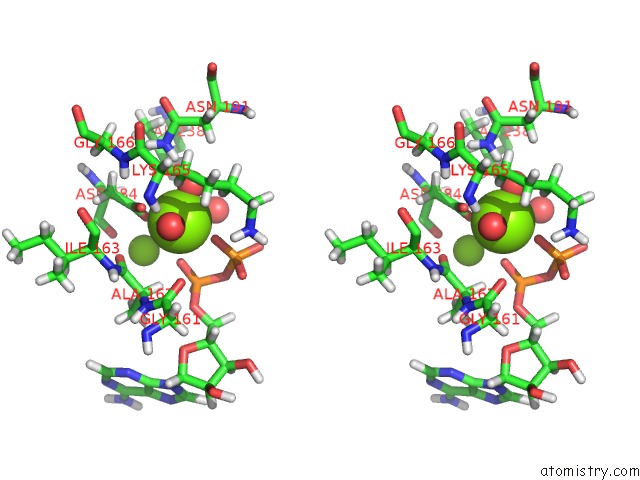
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Crystal Structure of Treponema Pallidum TP0796 Flavin Trafficking Protein, Adp Inhibited Form within 5.0Å range:
|
Reference:
R.K.Deka,
C.A.Brautigam,
W.Z.Liu,
D.R.Tomchick,
M.V.Norgard.
The TP0796 Lipoprotein of Treponema Pallidum Is A Bimetal-Dependent Fad Pyrophosphatase with A Potential Role in Flavin Homeostasis. J.Biol.Chem. V. 288 11106 2013.
ISSN: ISSN 0021-9258
PubMed: 23447540
DOI: 10.1074/JBC.M113.449975
Page generated: Fri Aug 16 16:42:05 2024
ISSN: ISSN 0021-9258
PubMed: 23447540
DOI: 10.1074/JBC.M113.449975
Last articles
Ca in 5UQZCa in 5ULY
Ca in 5UP7
Ca in 5UN3
Ca in 5UO9
Ca in 5ULU
Ca in 5UKG
Ca in 5UMH
Ca in 5UM1
Ca in 5ULV