Magnesium »
PDB 4jju-4jty »
4jr7 »
Magnesium in PDB 4jr7: Crystal Structure of SCCK2 Alpha in Complex with Gmppnp
Enzymatic activity of Crystal Structure of SCCK2 Alpha in Complex with Gmppnp
All present enzymatic activity of Crystal Structure of SCCK2 Alpha in Complex with Gmppnp:
2.7.11.1;
2.7.11.1;
Protein crystallography data
The structure of Crystal Structure of SCCK2 Alpha in Complex with Gmppnp, PDB code: 4jr7
was solved by
H.Liu,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 36.49 / 1.48 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 57.745, 69.512, 94.032, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 18.6 / 20.4 |
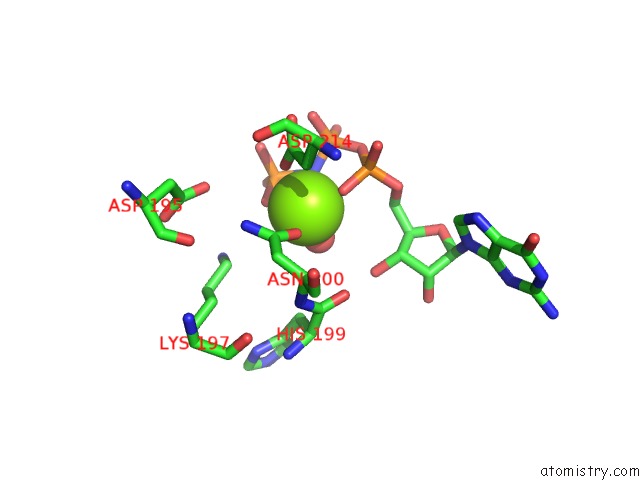
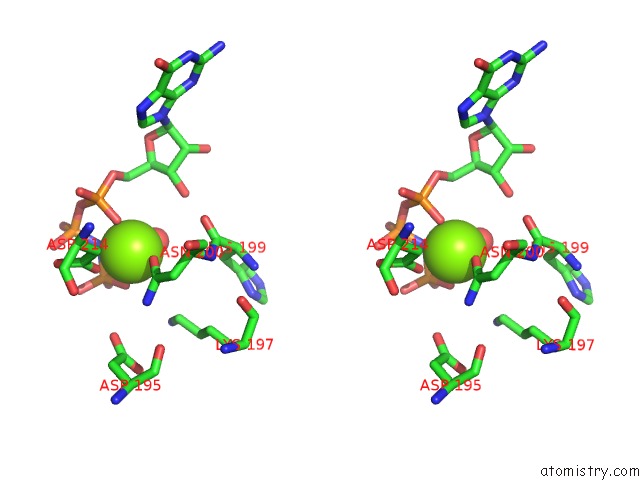
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Crystal Structure of SCCK2 Alpha in Complex with Gmppnp
(pdb code 4jr7). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total only one binding site of Magnesium was determined in the Crystal Structure of SCCK2 Alpha in Complex with Gmppnp, PDB code: 4jr7:
In total only one binding site of Magnesium was determined in the Crystal Structure of SCCK2 Alpha in Complex with Gmppnp, PDB code: 4jr7:
Magnesium binding site 1 out of 1 in 4jr7
Go back to
Magnesium binding site 1 out
of 1 in the Crystal Structure of SCCK2 Alpha in Complex with Gmppnp
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Crystal Structure of SCCK2 Alpha in Complex with Gmppnp within 5.0Å range:
|
Reference:
H.Liu,
H.Wang,
M.Teng,
X.Li.
The Multiple Nucleotide-Divalent Cation Binding Modes of Saccharomyces Cerevisiae CK2 Alpha Indicate A Possible Co-Substrate Hydrolysis Product (Adp/Gdp) Release Pathway. Acta Crystallogr.,Sect.D V. 70 501 2014.
ISSN: ISSN 0907-4449
PubMed: 24531484
DOI: 10.1107/S1399004713027879
Page generated: Mon Aug 11 17:16:45 2025
ISSN: ISSN 0907-4449
PubMed: 24531484
DOI: 10.1107/S1399004713027879
Last articles
Mg in 5B48Mg in 5B47
Mg in 5B46
Mg in 5B30
Mg in 5B2Z
Mg in 5B2T
Mg in 5B2S
Mg in 5B2R
Mg in 5B25
Mg in 5AYR