Magnesium »
PDB 4jju-4jty »
4jrn »
Magnesium in PDB 4jrn: ROP18 Kinase Domain in Complex with Amp-Pnp and Sucrose
Protein crystallography data
The structure of ROP18 Kinase Domain in Complex with Amp-Pnp and Sucrose, PDB code: 4jrn
was solved by
D.Lim,
D.A.Gold,
J.Lindsay,
E.E.Rosowski,
W.Niedelman,
M.B.Yaffe,
J.P.J.Saeij,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 36.55 / 2.71 |
| Space group | P 31 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 48.650, 48.650, 293.746, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 23.2 / 28.9 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the ROP18 Kinase Domain in Complex with Amp-Pnp and Sucrose
(pdb code 4jrn). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 2 binding sites of Magnesium where determined in the ROP18 Kinase Domain in Complex with Amp-Pnp and Sucrose, PDB code: 4jrn:
Jump to Magnesium binding site number: 1; 2;
In total 2 binding sites of Magnesium where determined in the ROP18 Kinase Domain in Complex with Amp-Pnp and Sucrose, PDB code: 4jrn:
Jump to Magnesium binding site number: 1; 2;
Magnesium binding site 1 out of 2 in 4jrn
Go back to
Magnesium binding site 1 out
of 2 in the ROP18 Kinase Domain in Complex with Amp-Pnp and Sucrose
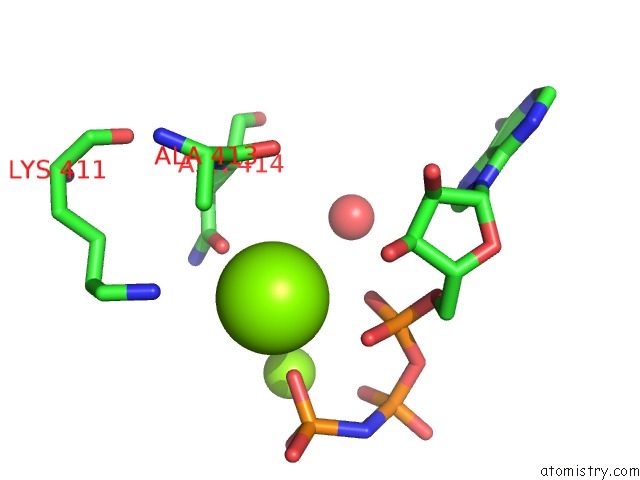
Mono view
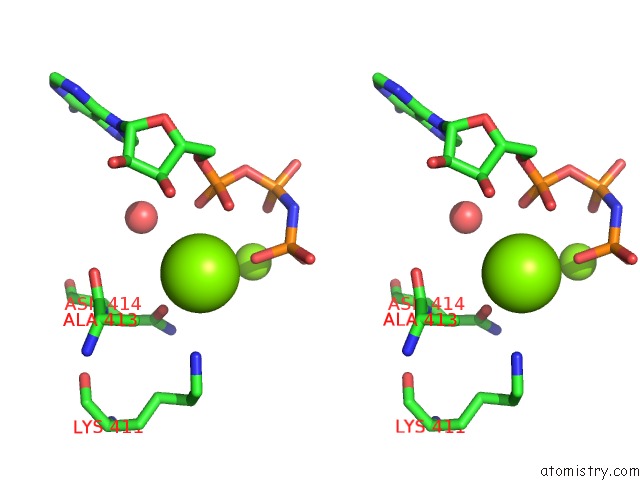
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of ROP18 Kinase Domain in Complex with Amp-Pnp and Sucrose within 5.0Å range:
|
Magnesium binding site 2 out of 2 in 4jrn
Go back to
Magnesium binding site 2 out
of 2 in the ROP18 Kinase Domain in Complex with Amp-Pnp and Sucrose
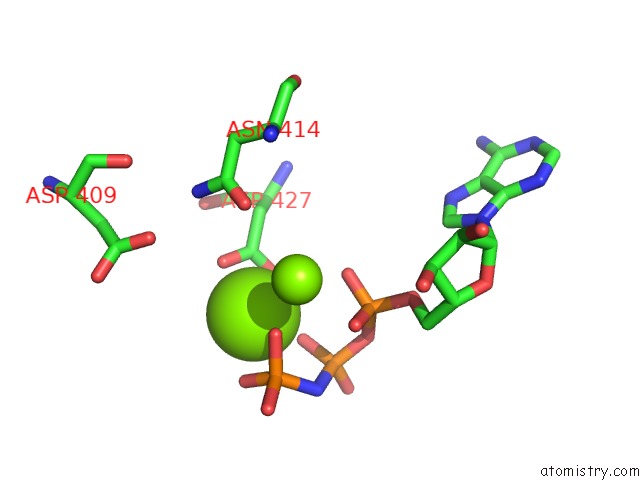
Mono view
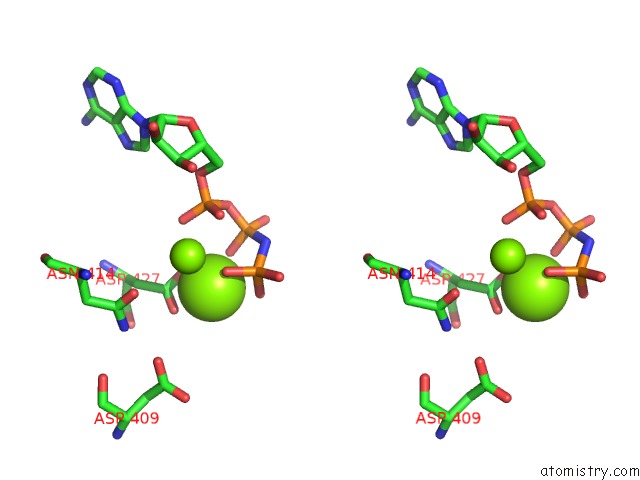
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of ROP18 Kinase Domain in Complex with Amp-Pnp and Sucrose within 5.0Å range:
|
Reference:
D.Lim,
D.A.Gold,
L.Julien,
E.E.Rosowski,
W.Niedelman,
M.B.Yaffe,
J.P.Saeij.
Structure of the Toxoplasma Gondii ROP18 Kinase Domain Reveals A Second Ligand Binding Pocket Required For Acute Virulence. J.Biol.Chem. V. 288 34968 2013.
ISSN: ISSN 0021-9258
PubMed: 24129568
DOI: 10.1074/JBC.M113.523266
Page generated: Mon Aug 11 17:16:51 2025
ISSN: ISSN 0021-9258
PubMed: 24129568
DOI: 10.1074/JBC.M113.523266
Last articles
Mg in 4RHDMg in 4RH7
Mg in 4RET
Mg in 4RGV
Mg in 4RGF
Mg in 4RF5
Mg in 4RF4
Mg in 4RG8
Mg in 4RES
Mg in 4RF2