Magnesium »
PDB 4lja-4lsh »
4lni »
Magnesium in PDB 4lni: B. Subtilis Glutamine Synthetase Structures Reveal Large Active Site Conformational Changes and Basis For Isoenzyme Specific Regulation: Structure of the Transition State Complex
Enzymatic activity of B. Subtilis Glutamine Synthetase Structures Reveal Large Active Site Conformational Changes and Basis For Isoenzyme Specific Regulation: Structure of the Transition State Complex
All present enzymatic activity of B. Subtilis Glutamine Synthetase Structures Reveal Large Active Site Conformational Changes and Basis For Isoenzyme Specific Regulation: Structure of the Transition State Complex:
6.3.1.2;
6.3.1.2;
Protein crystallography data
The structure of B. Subtilis Glutamine Synthetase Structures Reveal Large Active Site Conformational Changes and Basis For Isoenzyme Specific Regulation: Structure of the Transition State Complex, PDB code: 4lni
was solved by
M.A.Schumacher,
N.Chinnam,
N.Tonthat,
S.Fisher,
L.Wray,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 117.10 / 2.58 |
| Space group | P 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 110.200, 141.600, 142.100, 60.29, 67.38, 76.20 |
| R / Rfree (%) | 16.5 / 22.3 |
Magnesium Binding Sites:
Pages:
>>> Page 1 <<< Page 2, Binding sites: 11 - 20; Page 3, Binding sites: 21 - 30; Page 4, Binding sites: 31 - 36;Binding sites:
The binding sites of Magnesium atom in the B. Subtilis Glutamine Synthetase Structures Reveal Large Active Site Conformational Changes and Basis For Isoenzyme Specific Regulation: Structure of the Transition State Complex (pdb code 4lni). This binding sites where shown within 5.0 Angstroms radius around Magnesium atom.In total 36 binding sites of Magnesium where determined in the B. Subtilis Glutamine Synthetase Structures Reveal Large Active Site Conformational Changes and Basis For Isoenzyme Specific Regulation: Structure of the Transition State Complex, PDB code: 4lni:
Jump to Magnesium binding site number: 1; 2; 3; 4; 5; 6; 7; 8; 9; 10;
Magnesium binding site 1 out of 36 in 4lni
Go back to
Magnesium binding site 1 out
of 36 in the B. Subtilis Glutamine Synthetase Structures Reveal Large Active Site Conformational Changes and Basis For Isoenzyme Specific Regulation: Structure of the Transition State Complex
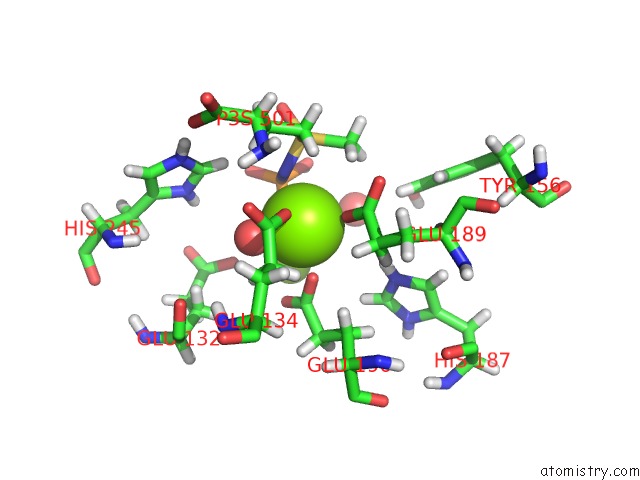
Mono view
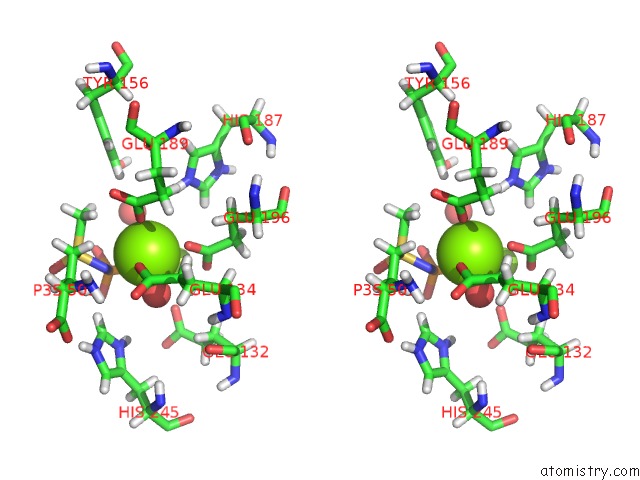
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of B. Subtilis Glutamine Synthetase Structures Reveal Large Active Site Conformational Changes and Basis For Isoenzyme Specific Regulation: Structure of the Transition State Complex within 5.0Å range:
|
Magnesium binding site 2 out of 36 in 4lni
Go back to
Magnesium binding site 2 out
of 36 in the B. Subtilis Glutamine Synthetase Structures Reveal Large Active Site Conformational Changes and Basis For Isoenzyme Specific Regulation: Structure of the Transition State Complex
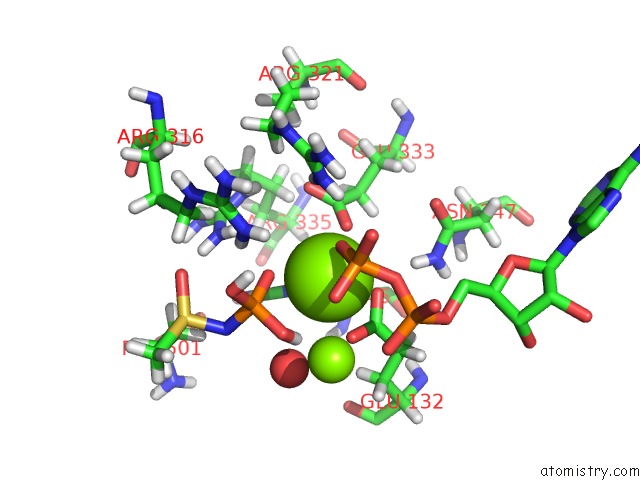
Mono view
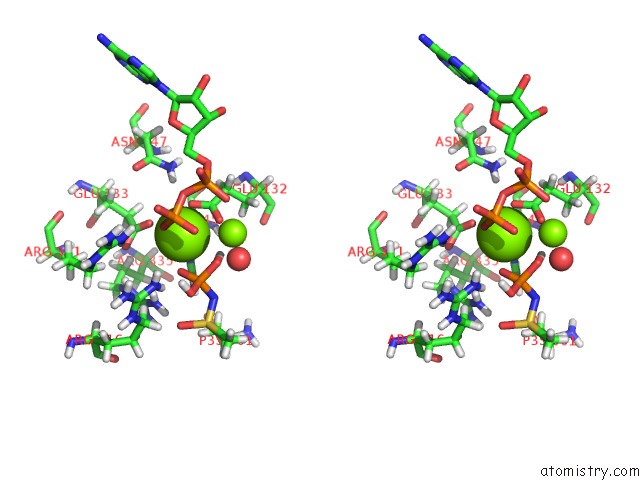
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of B. Subtilis Glutamine Synthetase Structures Reveal Large Active Site Conformational Changes and Basis For Isoenzyme Specific Regulation: Structure of the Transition State Complex within 5.0Å range:
|
Magnesium binding site 3 out of 36 in 4lni
Go back to
Magnesium binding site 3 out
of 36 in the B. Subtilis Glutamine Synthetase Structures Reveal Large Active Site Conformational Changes and Basis For Isoenzyme Specific Regulation: Structure of the Transition State Complex
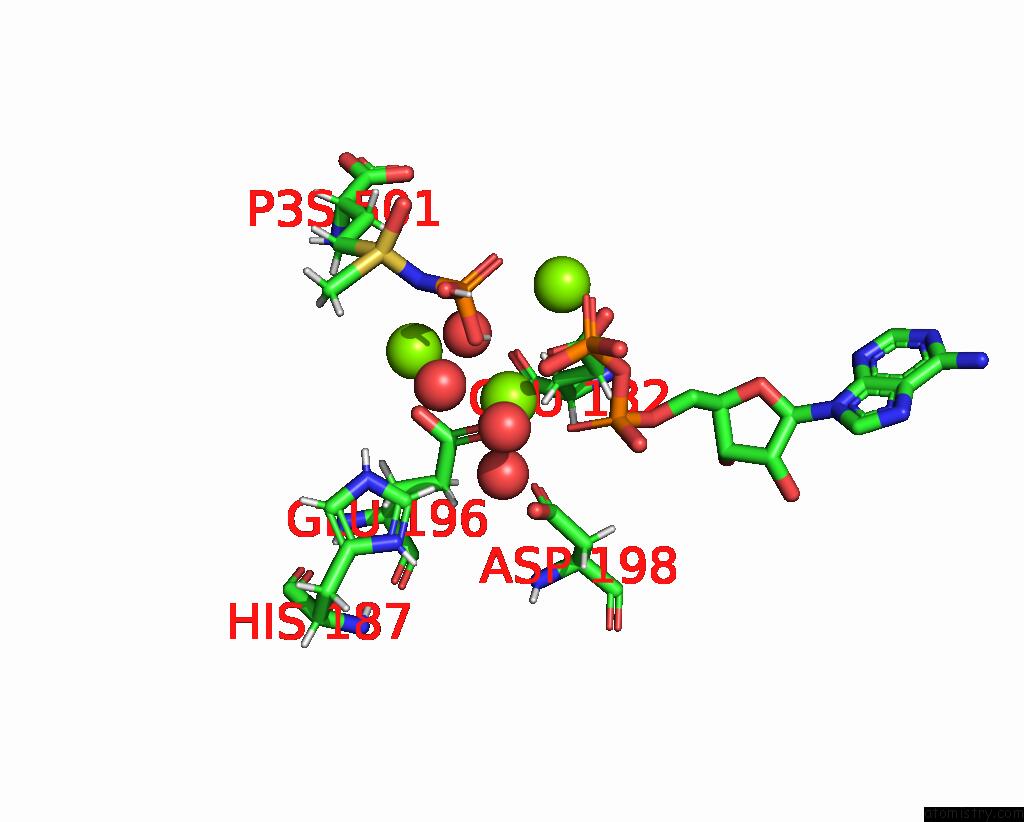
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 3 of B. Subtilis Glutamine Synthetase Structures Reveal Large Active Site Conformational Changes and Basis For Isoenzyme Specific Regulation: Structure of the Transition State Complex within 5.0Å range:
|
Magnesium binding site 4 out of 36 in 4lni
Go back to
Magnesium binding site 4 out
of 36 in the B. Subtilis Glutamine Synthetase Structures Reveal Large Active Site Conformational Changes and Basis For Isoenzyme Specific Regulation: Structure of the Transition State Complex
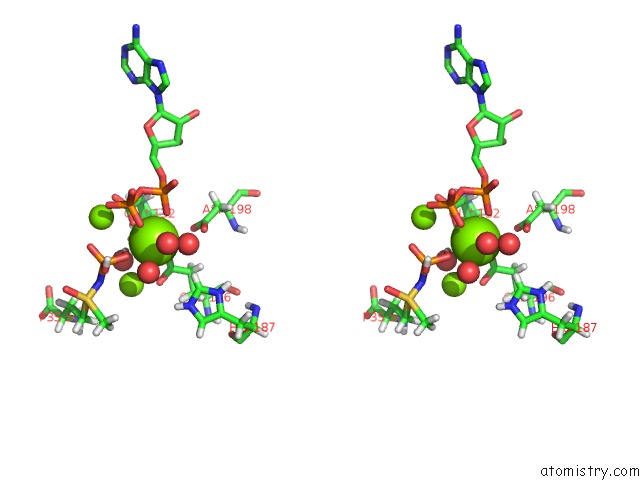
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 4 of B. Subtilis Glutamine Synthetase Structures Reveal Large Active Site Conformational Changes and Basis For Isoenzyme Specific Regulation: Structure of the Transition State Complex within 5.0Å range:
|
Magnesium binding site 5 out of 36 in 4lni
Go back to
Magnesium binding site 5 out
of 36 in the B. Subtilis Glutamine Synthetase Structures Reveal Large Active Site Conformational Changes and Basis For Isoenzyme Specific Regulation: Structure of the Transition State Complex
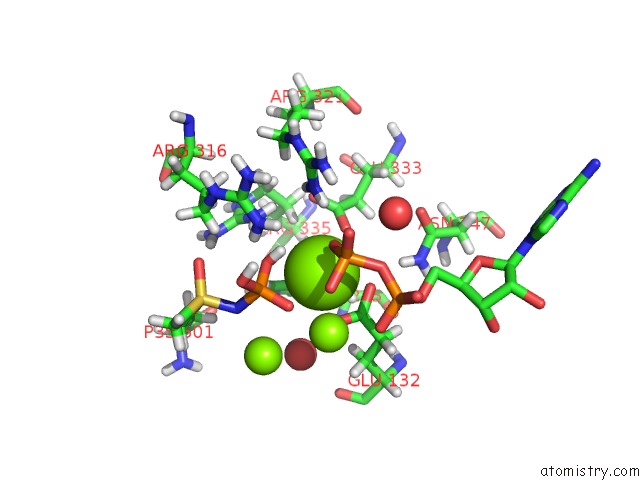
Mono view
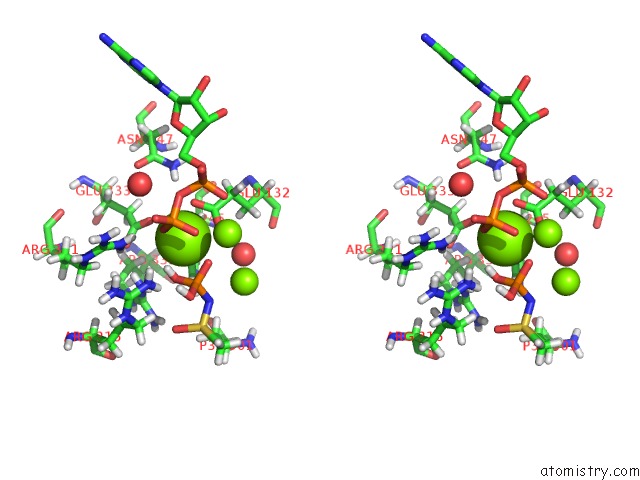
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 5 of B. Subtilis Glutamine Synthetase Structures Reveal Large Active Site Conformational Changes and Basis For Isoenzyme Specific Regulation: Structure of the Transition State Complex within 5.0Å range:
|
Magnesium binding site 6 out of 36 in 4lni
Go back to
Magnesium binding site 6 out
of 36 in the B. Subtilis Glutamine Synthetase Structures Reveal Large Active Site Conformational Changes and Basis For Isoenzyme Specific Regulation: Structure of the Transition State Complex
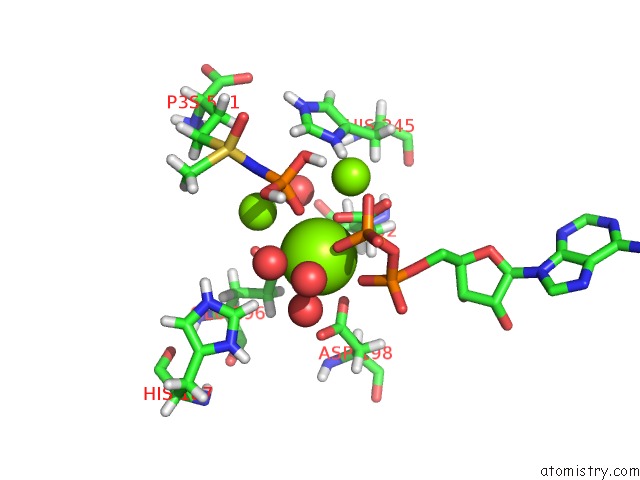
Mono view
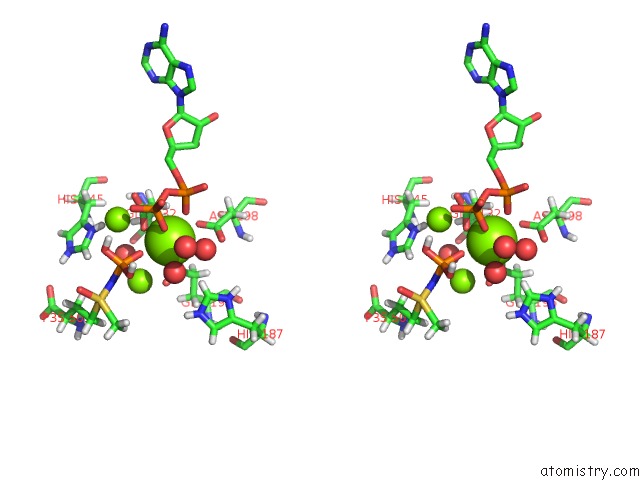
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 6 of B. Subtilis Glutamine Synthetase Structures Reveal Large Active Site Conformational Changes and Basis For Isoenzyme Specific Regulation: Structure of the Transition State Complex within 5.0Å range:
|
Magnesium binding site 7 out of 36 in 4lni
Go back to
Magnesium binding site 7 out
of 36 in the B. Subtilis Glutamine Synthetase Structures Reveal Large Active Site Conformational Changes and Basis For Isoenzyme Specific Regulation: Structure of the Transition State Complex
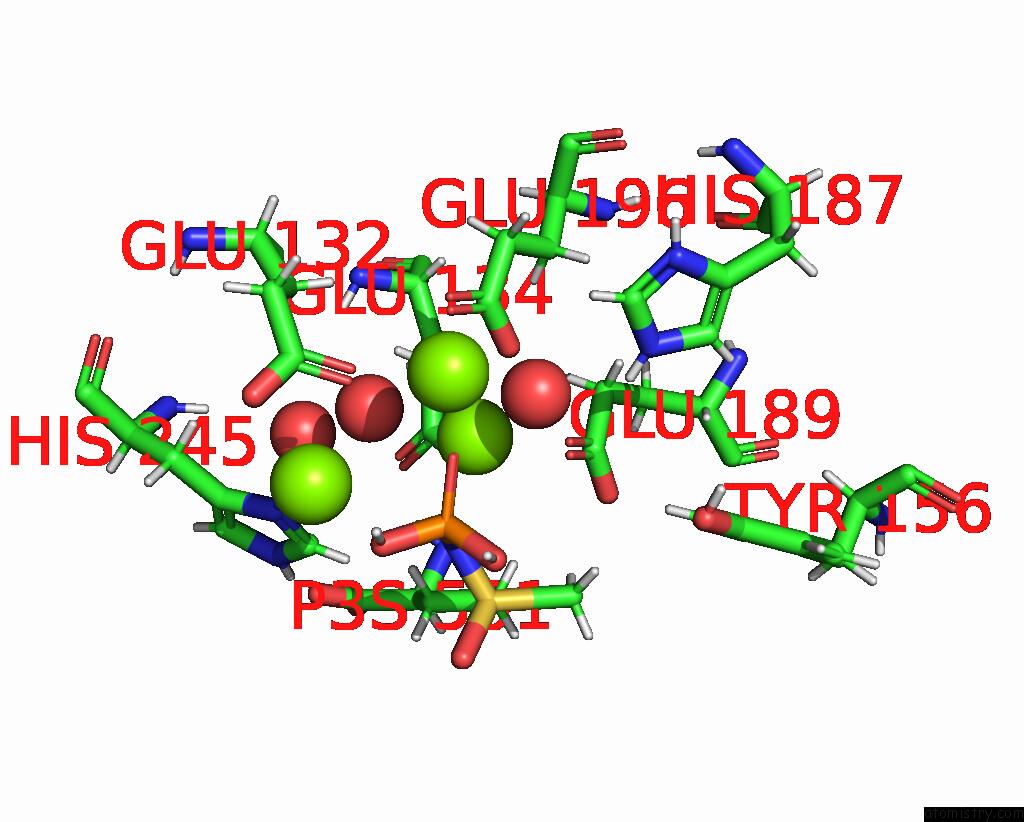
Mono view
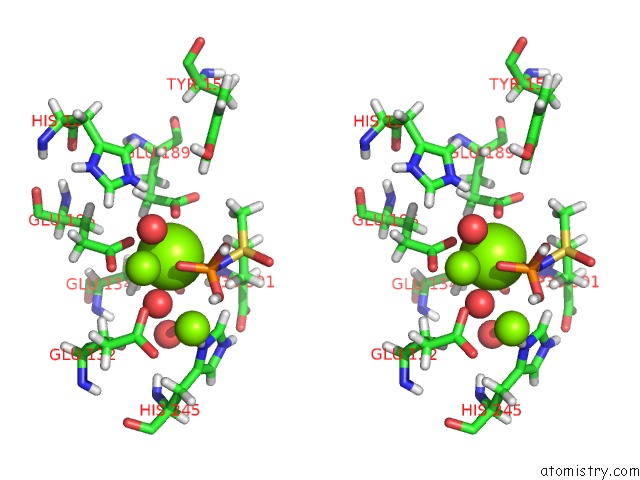
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 7 of B. Subtilis Glutamine Synthetase Structures Reveal Large Active Site Conformational Changes and Basis For Isoenzyme Specific Regulation: Structure of the Transition State Complex within 5.0Å range:
|
Magnesium binding site 8 out of 36 in 4lni
Go back to
Magnesium binding site 8 out
of 36 in the B. Subtilis Glutamine Synthetase Structures Reveal Large Active Site Conformational Changes and Basis For Isoenzyme Specific Regulation: Structure of the Transition State Complex
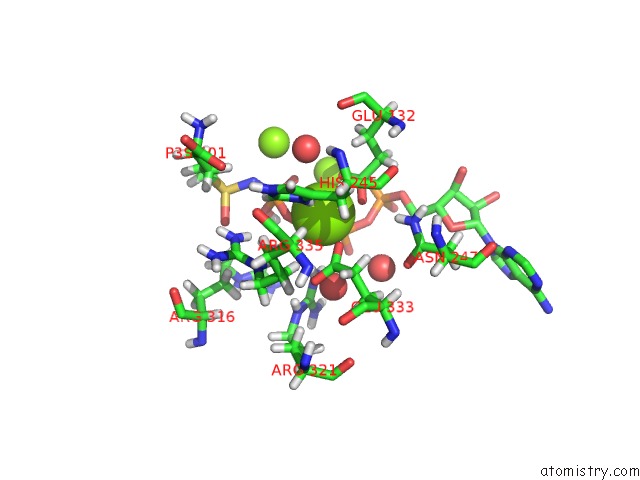
Mono view
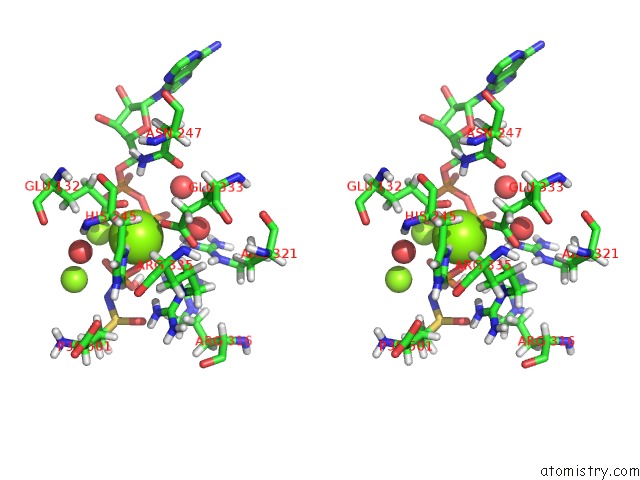
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 8 of B. Subtilis Glutamine Synthetase Structures Reveal Large Active Site Conformational Changes and Basis For Isoenzyme Specific Regulation: Structure of the Transition State Complex within 5.0Å range:
|
Magnesium binding site 9 out of 36 in 4lni
Go back to
Magnesium binding site 9 out
of 36 in the B. Subtilis Glutamine Synthetase Structures Reveal Large Active Site Conformational Changes and Basis For Isoenzyme Specific Regulation: Structure of the Transition State Complex
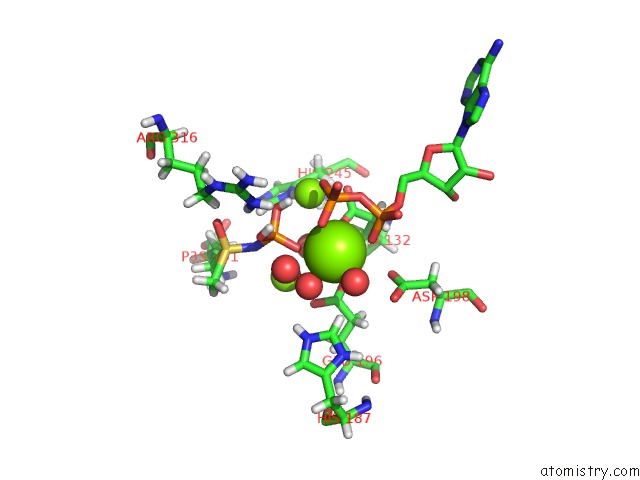
Mono view
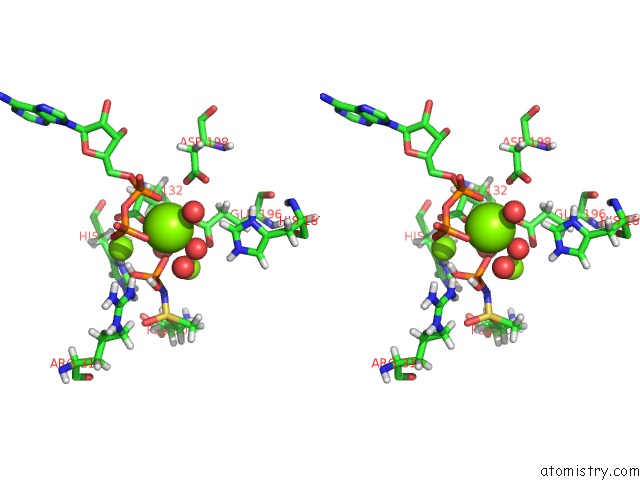
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 9 of B. Subtilis Glutamine Synthetase Structures Reveal Large Active Site Conformational Changes and Basis For Isoenzyme Specific Regulation: Structure of the Transition State Complex within 5.0Å range:
|
Magnesium binding site 10 out of 36 in 4lni
Go back to
Magnesium binding site 10 out
of 36 in the B. Subtilis Glutamine Synthetase Structures Reveal Large Active Site Conformational Changes and Basis For Isoenzyme Specific Regulation: Structure of the Transition State Complex
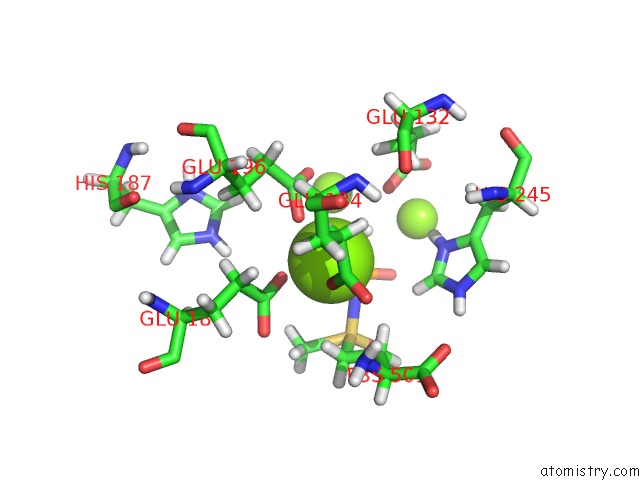
Mono view
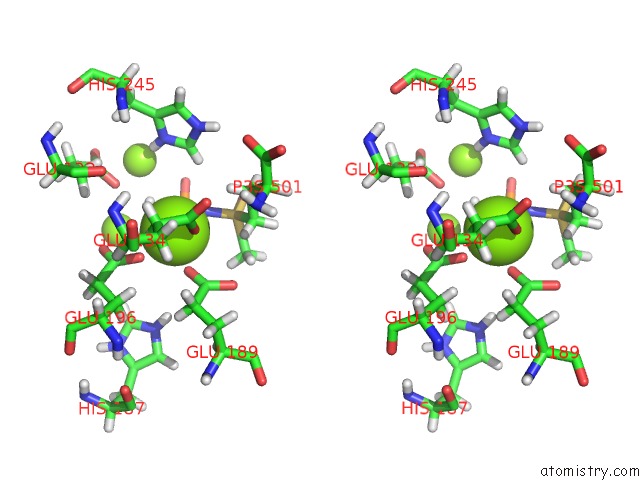
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 10 of B. Subtilis Glutamine Synthetase Structures Reveal Large Active Site Conformational Changes and Basis For Isoenzyme Specific Regulation: Structure of the Transition State Complex within 5.0Å range:
|
Reference:
D.S.Murray,
N.Chinnam,
N.K.Tonthat,
T.Whitfill,
L.V.Wray,
S.H.Fisher,
M.A.Schumacher.
Structures of the Bacillus Subtilis Glutamine Synthetase Dodecamer Reveal Large Intersubunit Catalytic Conformational Changes Linked to A Unique Feedback Inhibition Mechanism. J.Biol.Chem. V. 288 35801 2013.
ISSN: ISSN 0021-9258
PubMed: 24158439
DOI: 10.1074/JBC.M113.519496
Page generated: Mon Aug 11 19:55:49 2025
ISSN: ISSN 0021-9258
PubMed: 24158439
DOI: 10.1074/JBC.M113.519496
Last articles
Mg in 4QZ3Mg in 4QZ2
Mg in 4QZ0
Mg in 4QZ1
Mg in 4QYI
Mg in 4QYT
Mg in 4QYM
Mg in 4QY6
Mg in 4QXJ
Mg in 4QY5