Magnesium »
PDB 4lja-4lsh »
4lrw »
Magnesium in PDB 4lrw: Crystal Structure of K-Ras G12C (Cysteine-Light), Gdp-Bound
Protein crystallography data
The structure of Crystal Structure of K-Ras G12C (Cysteine-Light), Gdp-Bound, PDB code: 4lrw
was solved by
J.M.Ostrem,
U.Peters,
M.L.Sos,
J.A.Wells,
K.M.Shokat,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 23.92 / 2.15 |
| Space group | P 3 |
| Cell size a, b, c (Å), α, β, γ (°) | 82.862, 82.862, 38.397, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 18 / 20.4 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Crystal Structure of K-Ras G12C (Cysteine-Light), Gdp-Bound
(pdb code 4lrw). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 4 binding sites of Magnesium where determined in the Crystal Structure of K-Ras G12C (Cysteine-Light), Gdp-Bound, PDB code: 4lrw:
Jump to Magnesium binding site number: 1; 2; 3; 4;
In total 4 binding sites of Magnesium where determined in the Crystal Structure of K-Ras G12C (Cysteine-Light), Gdp-Bound, PDB code: 4lrw:
Jump to Magnesium binding site number: 1; 2; 3; 4;
Magnesium binding site 1 out of 4 in 4lrw
Go back to
Magnesium binding site 1 out
of 4 in the Crystal Structure of K-Ras G12C (Cysteine-Light), Gdp-Bound
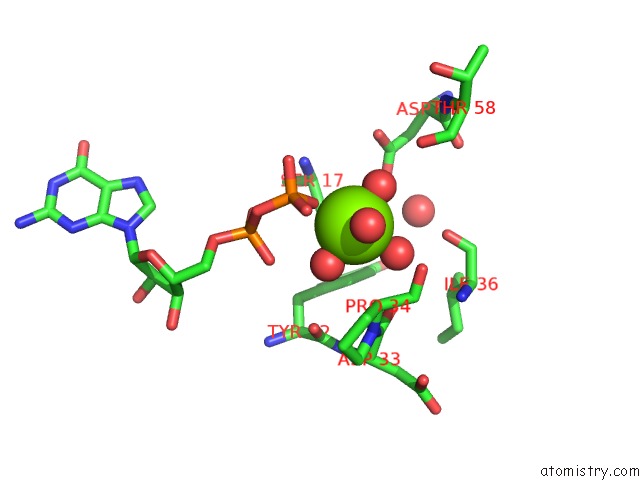
Mono view
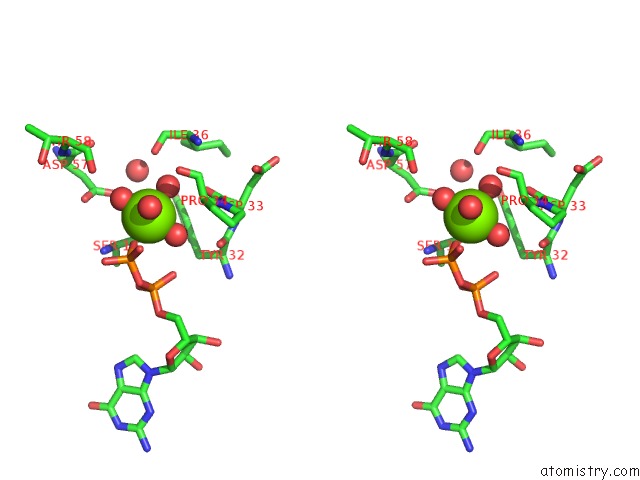
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Crystal Structure of K-Ras G12C (Cysteine-Light), Gdp-Bound within 5.0Å range:
|
Magnesium binding site 2 out of 4 in 4lrw
Go back to
Magnesium binding site 2 out
of 4 in the Crystal Structure of K-Ras G12C (Cysteine-Light), Gdp-Bound
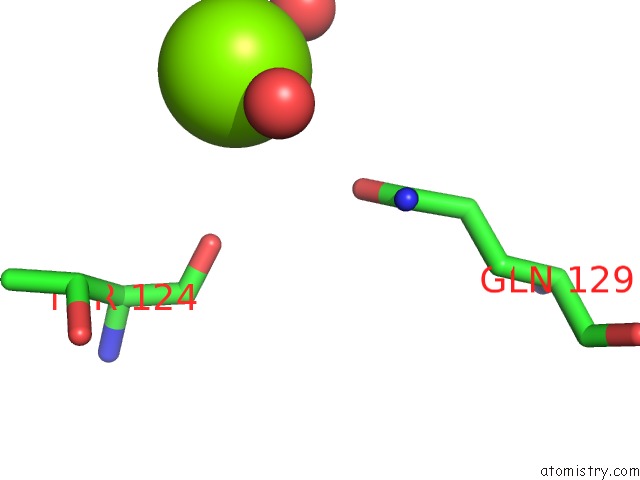
Mono view
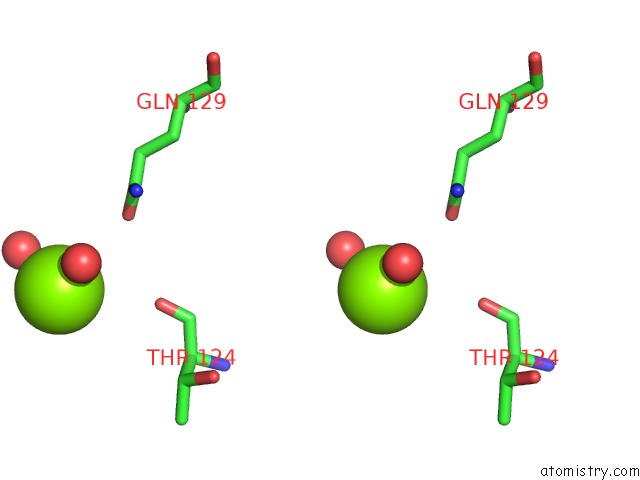
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Crystal Structure of K-Ras G12C (Cysteine-Light), Gdp-Bound within 5.0Å range:
|
Magnesium binding site 3 out of 4 in 4lrw
Go back to
Magnesium binding site 3 out
of 4 in the Crystal Structure of K-Ras G12C (Cysteine-Light), Gdp-Bound
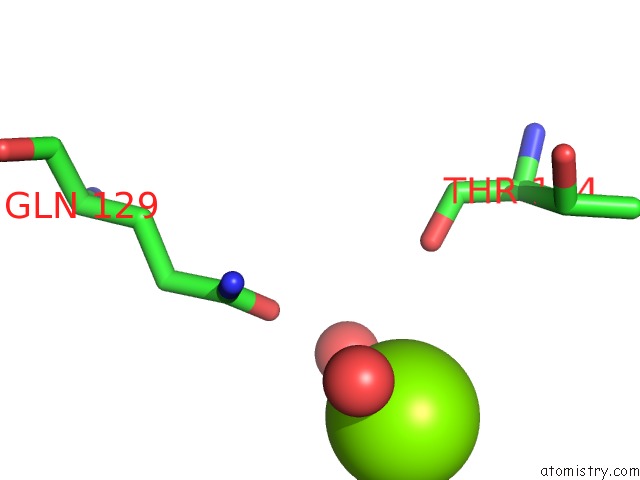
Mono view
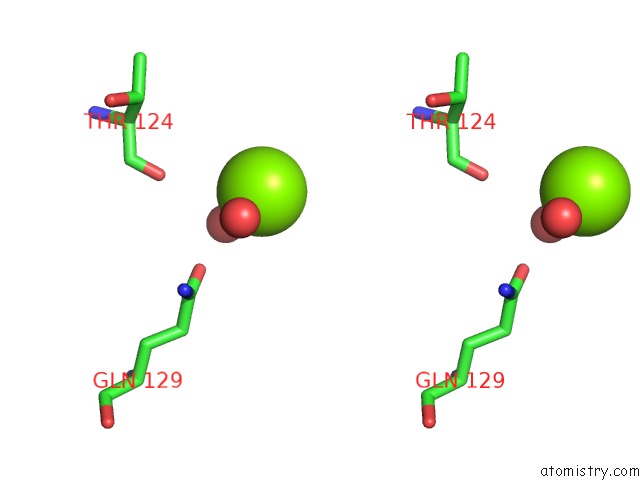
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 3 of Crystal Structure of K-Ras G12C (Cysteine-Light), Gdp-Bound within 5.0Å range:
|
Magnesium binding site 4 out of 4 in 4lrw
Go back to
Magnesium binding site 4 out
of 4 in the Crystal Structure of K-Ras G12C (Cysteine-Light), Gdp-Bound
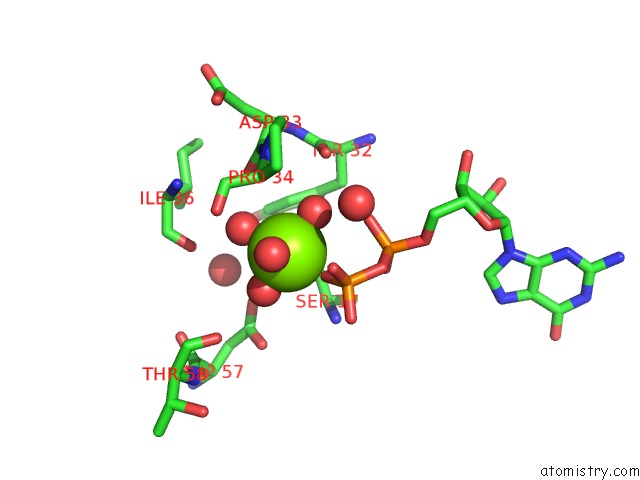
Mono view
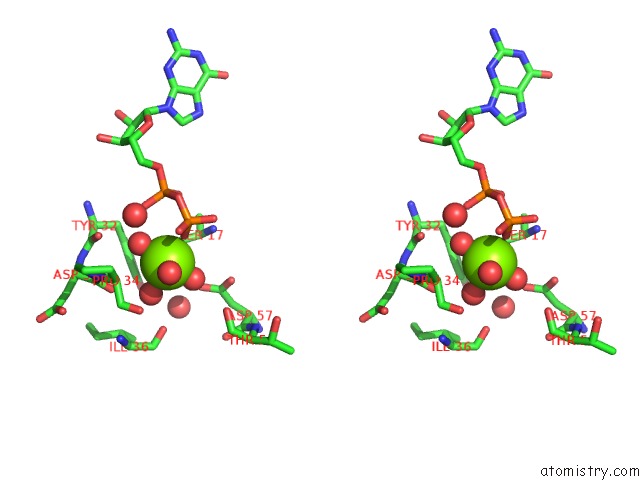
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 4 of Crystal Structure of K-Ras G12C (Cysteine-Light), Gdp-Bound within 5.0Å range:
|
Reference:
J.M.Ostrem,
U.Peters,
M.L.Sos,
J.A.Wells,
K.M.Shokat.
K-Ras(G12C) Inhibitors Allosterically Control Gtp Affinity and Effector Interactions. Nature V. 503 548 2013.
ISSN: ISSN 0028-0836
PubMed: 24256730
DOI: 10.1038/NATURE12796
Page generated: Mon Aug 11 20:07:27 2025
ISSN: ISSN 0028-0836
PubMed: 24256730
DOI: 10.1038/NATURE12796
Last articles
Mg in 4RKDMg in 4RKF
Mg in 4RJJ
Mg in 4RKE
Mg in 4RKC
Mg in 4RGE
Mg in 4RJL
Mg in 4RJI
Mg in 4RIY
Mg in 4RIX