Magnesium »
PDB 4nbm-4nlz »
4ncf »
Magnesium in PDB 4ncf: Crystal Structure of Eukaryotic Translation Initiation Factor EIF5B (399-852) From Saccharomyces Cerevisiae in Complex with Gdp
Protein crystallography data
The structure of Crystal Structure of Eukaryotic Translation Initiation Factor EIF5B (399-852) From Saccharomyces Cerevisiae in Complex with Gdp, PDB code: 4ncf
was solved by
B.Kuhle,
R.Ficner,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 46.66 / 3.02 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 73.560, 119.460, 120.730, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 25.3 / 28.1 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Crystal Structure of Eukaryotic Translation Initiation Factor EIF5B (399-852) From Saccharomyces Cerevisiae in Complex with Gdp
(pdb code 4ncf). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 2 binding sites of Magnesium where determined in the Crystal Structure of Eukaryotic Translation Initiation Factor EIF5B (399-852) From Saccharomyces Cerevisiae in Complex with Gdp, PDB code: 4ncf:
Jump to Magnesium binding site number: 1; 2;
In total 2 binding sites of Magnesium where determined in the Crystal Structure of Eukaryotic Translation Initiation Factor EIF5B (399-852) From Saccharomyces Cerevisiae in Complex with Gdp, PDB code: 4ncf:
Jump to Magnesium binding site number: 1; 2;
Magnesium binding site 1 out of 2 in 4ncf
Go back to
Magnesium binding site 1 out
of 2 in the Crystal Structure of Eukaryotic Translation Initiation Factor EIF5B (399-852) From Saccharomyces Cerevisiae in Complex with Gdp
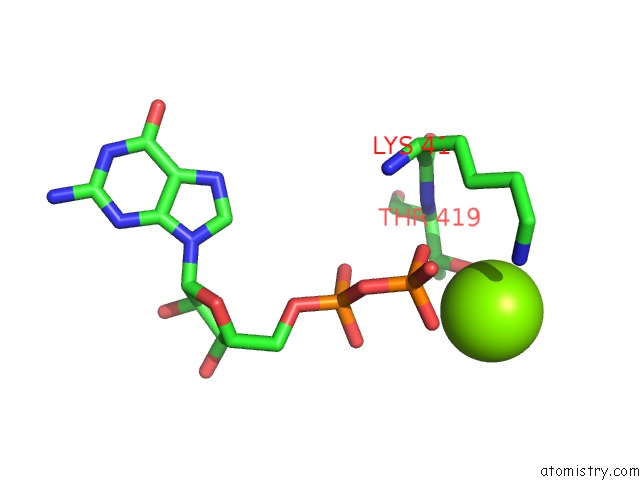
Mono view
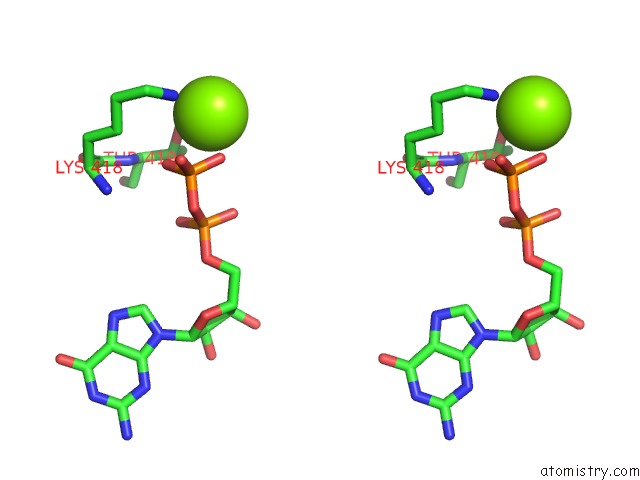
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Crystal Structure of Eukaryotic Translation Initiation Factor EIF5B (399-852) From Saccharomyces Cerevisiae in Complex with Gdp within 5.0Å range:
|
Magnesium binding site 2 out of 2 in 4ncf
Go back to
Magnesium binding site 2 out
of 2 in the Crystal Structure of Eukaryotic Translation Initiation Factor EIF5B (399-852) From Saccharomyces Cerevisiae in Complex with Gdp
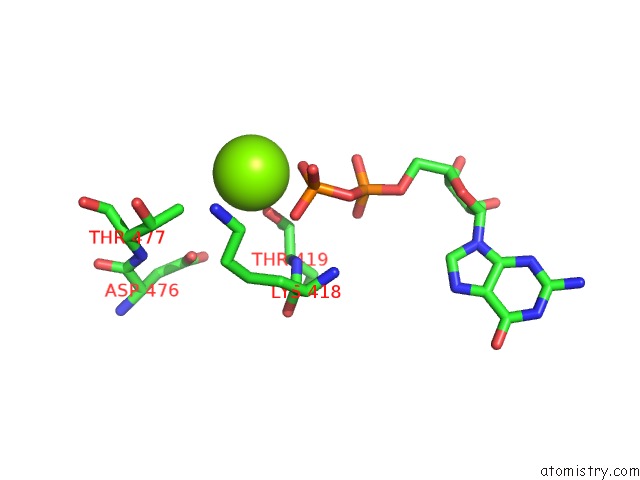
Mono view
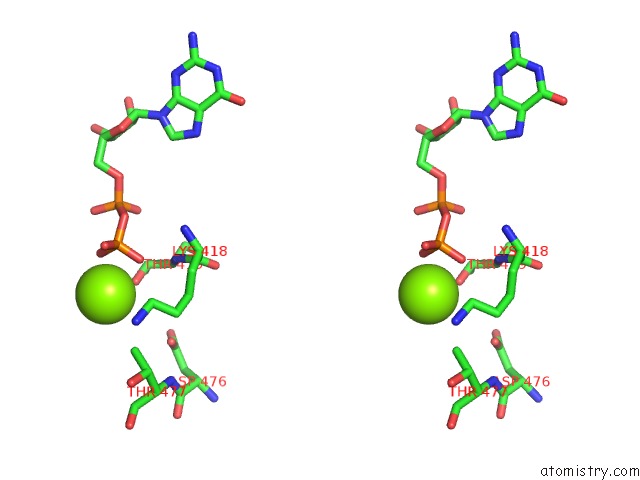
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Crystal Structure of Eukaryotic Translation Initiation Factor EIF5B (399-852) From Saccharomyces Cerevisiae in Complex with Gdp within 5.0Å range:
|
Reference:
B.Kuhle,
R.Ficner.
EIF5B Employs A Novel Domain Release Mechanism to Catalyze Ribosomal Subunit Joining. Embo J. V. 33 1177 2014.
ISSN: ISSN 0261-4189
PubMed: 24686316
DOI: 10.1002/EMBJ.201387344
Page generated: Mon Aug 11 20:40:03 2025
ISSN: ISSN 0261-4189
PubMed: 24686316
DOI: 10.1002/EMBJ.201387344
Last articles
Mg in 4UMWMg in 4UMV
Mg in 4UM7
Mg in 4UME
Mg in 4UM9
Mg in 4UM8
Mg in 4UB8
Mg in 4UB6
Mg in 4UM5
Mg in 4UKD