Magnesium »
PDB 4nbm-4nlz »
4ndn »
Magnesium in PDB 4ndn: Structural Insights of Mat Enzymes: MATA2B Complexed with Sam and Ppnp
Enzymatic activity of Structural Insights of Mat Enzymes: MATA2B Complexed with Sam and Ppnp
All present enzymatic activity of Structural Insights of Mat Enzymes: MATA2B Complexed with Sam and Ppnp:
2.5.1.6;
2.5.1.6;
Protein crystallography data
The structure of Structural Insights of Mat Enzymes: MATA2B Complexed with Sam and Ppnp, PDB code: 4ndn
was solved by
B.Murray,
S.V.Antonyuk,
A.Marina,
S.C.Lu,
J.M.Mato,
S.S.Hasnain,
A.L.Rojas,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 47.78 / 2.34 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 72.140, 122.178, 298.423, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 21.1 / 25.1 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Structural Insights of Mat Enzymes: MATA2B Complexed with Sam and Ppnp
(pdb code 4ndn). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 4 binding sites of Magnesium where determined in the Structural Insights of Mat Enzymes: MATA2B Complexed with Sam and Ppnp, PDB code: 4ndn:
Jump to Magnesium binding site number: 1; 2; 3; 4;
In total 4 binding sites of Magnesium where determined in the Structural Insights of Mat Enzymes: MATA2B Complexed with Sam and Ppnp, PDB code: 4ndn:
Jump to Magnesium binding site number: 1; 2; 3; 4;
Magnesium binding site 1 out of 4 in 4ndn
Go back to
Magnesium binding site 1 out
of 4 in the Structural Insights of Mat Enzymes: MATA2B Complexed with Sam and Ppnp
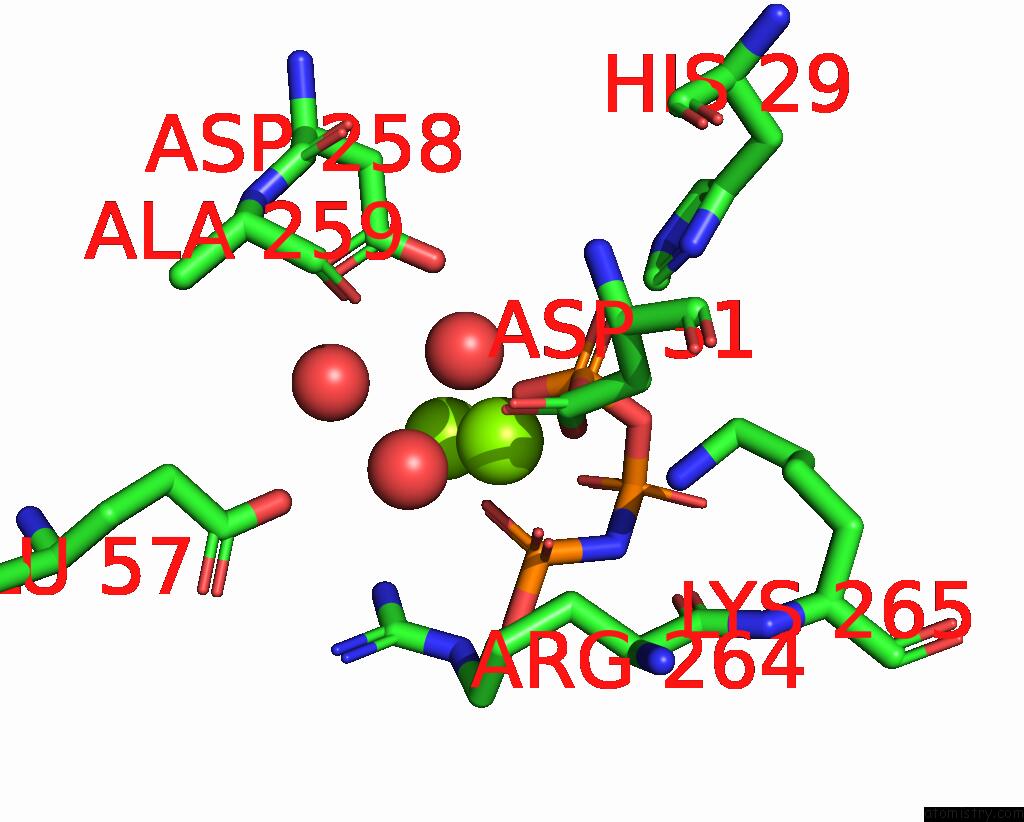
Mono view
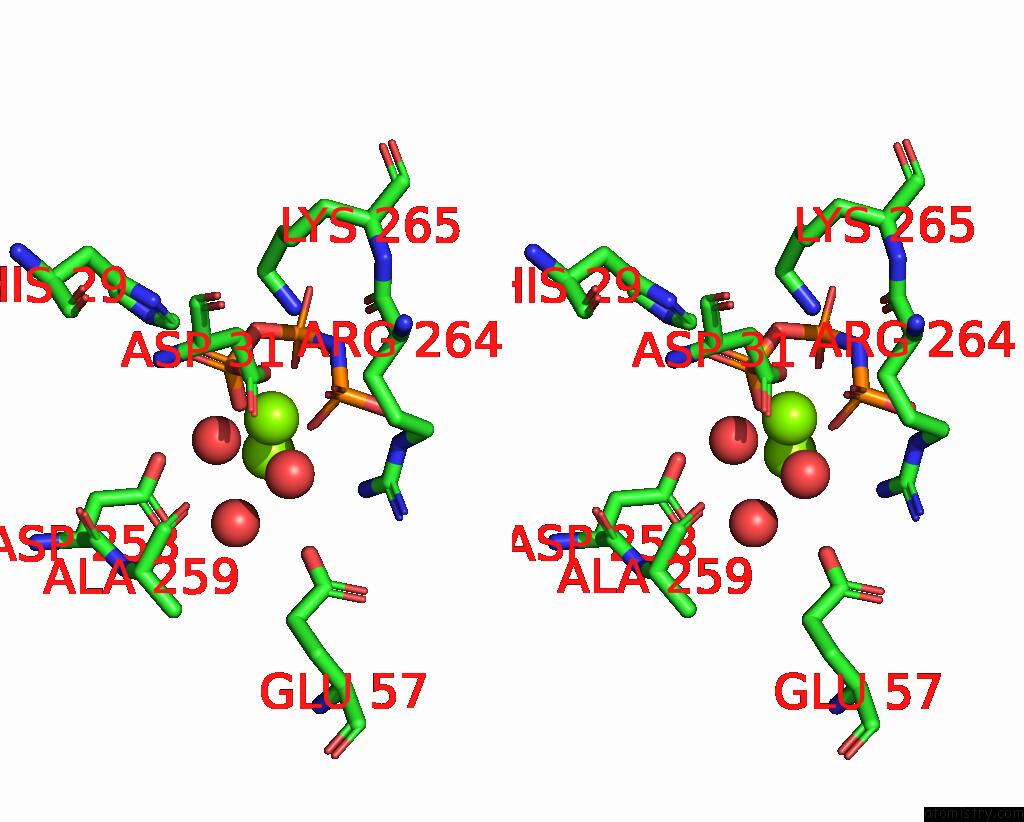
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Structural Insights of Mat Enzymes: MATA2B Complexed with Sam and Ppnp within 5.0Å range:
|
Magnesium binding site 2 out of 4 in 4ndn
Go back to
Magnesium binding site 2 out
of 4 in the Structural Insights of Mat Enzymes: MATA2B Complexed with Sam and Ppnp
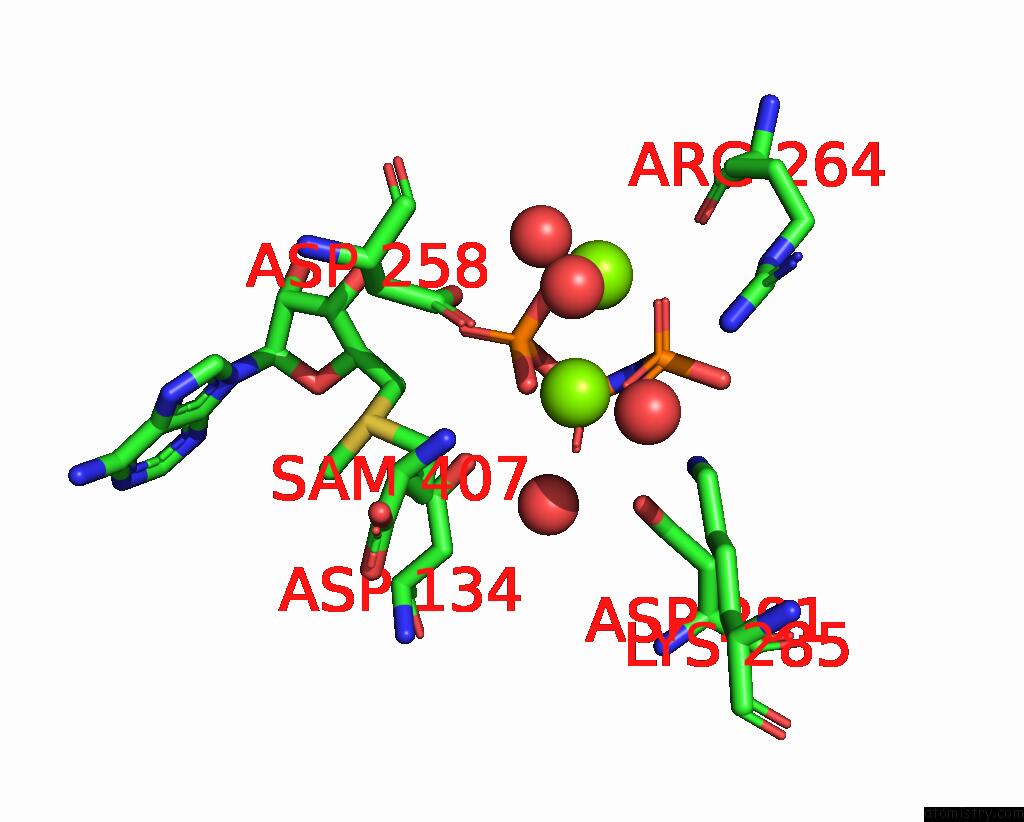
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Structural Insights of Mat Enzymes: MATA2B Complexed with Sam and Ppnp within 5.0Å range:
|
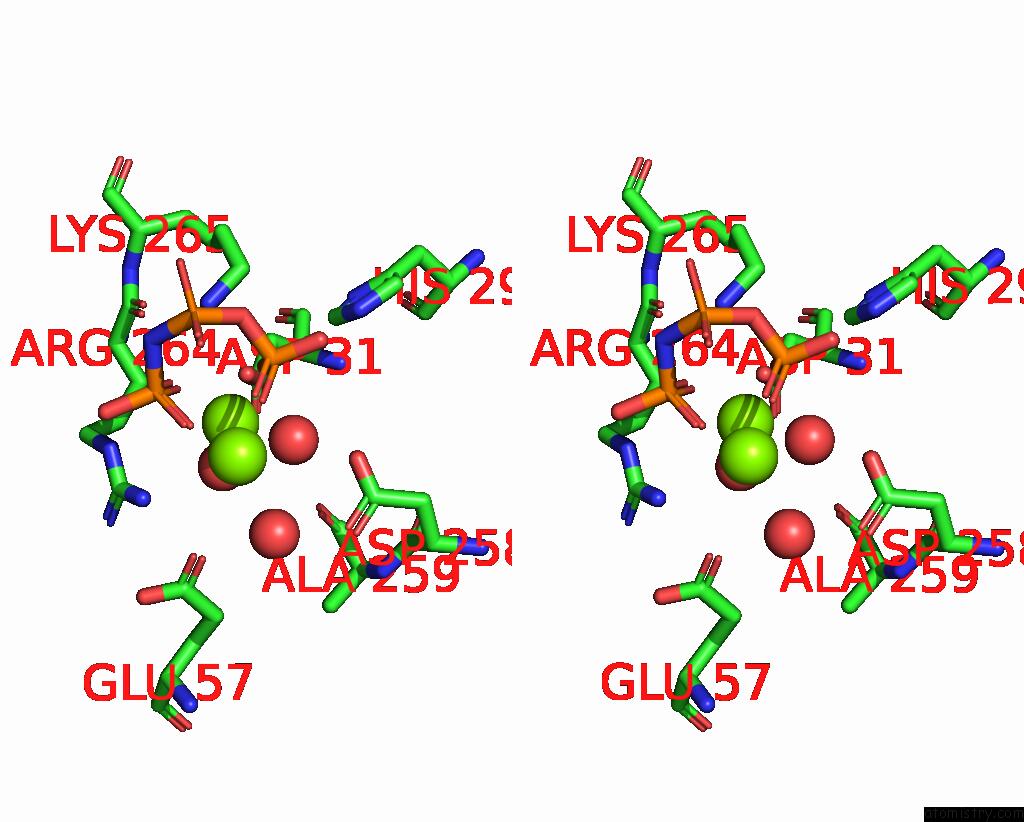
Magnesium binding site 3 out of 4 in 4ndn
Go back to
Magnesium binding site 3 out
of 4 in the Structural Insights of Mat Enzymes: MATA2B Complexed with Sam and Ppnp
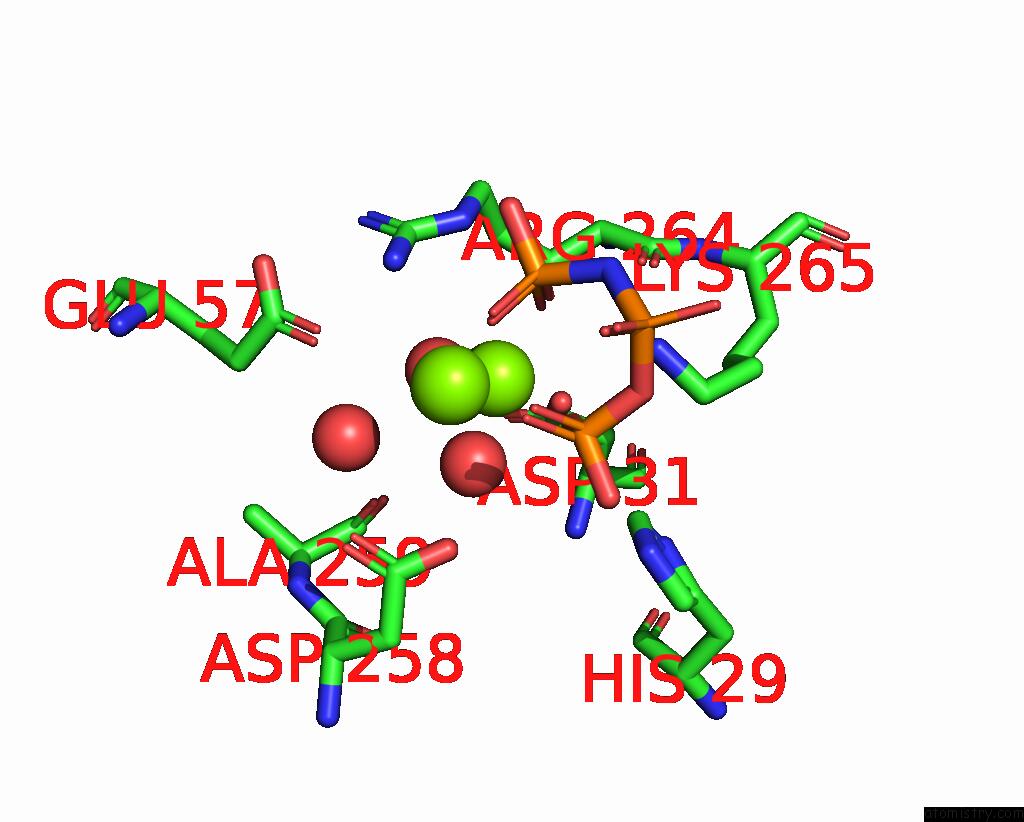
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 3 of Structural Insights of Mat Enzymes: MATA2B Complexed with Sam and Ppnp within 5.0Å range:
|
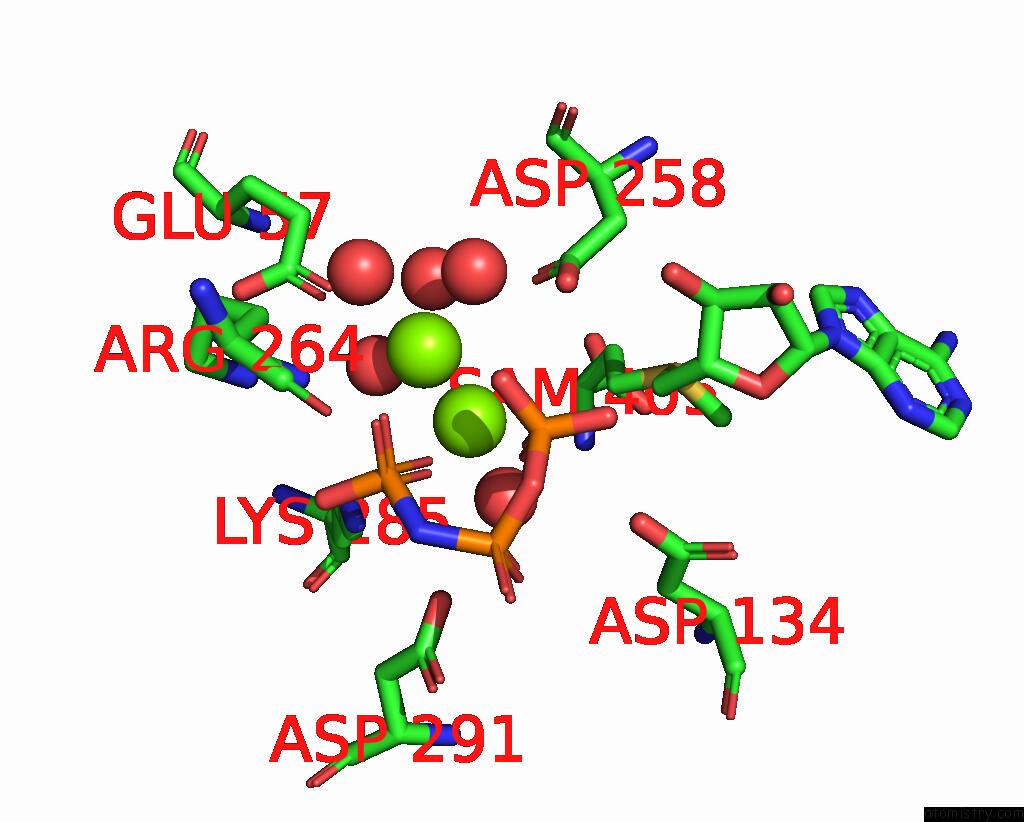
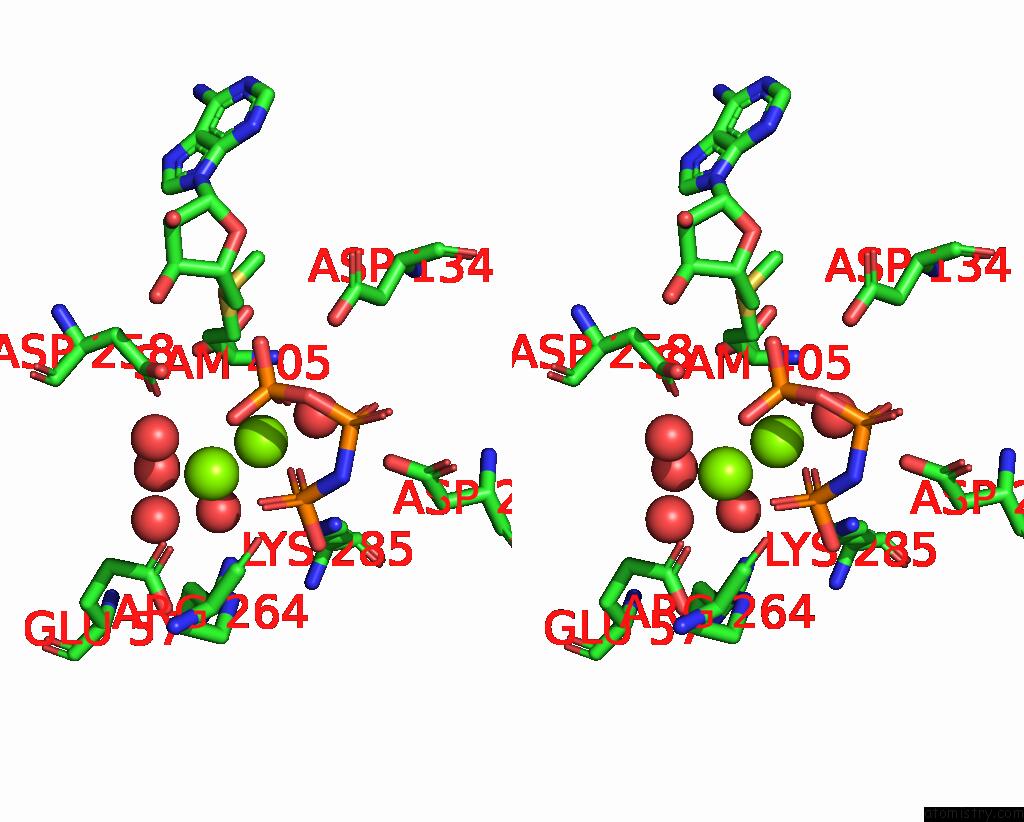
Magnesium binding site 4 out of 4 in 4ndn
Go back to
Magnesium binding site 4 out
of 4 in the Structural Insights of Mat Enzymes: MATA2B Complexed with Sam and Ppnp
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 4 of Structural Insights of Mat Enzymes: MATA2B Complexed with Sam and Ppnp within 5.0Å range:
|
Reference:
B.Murray,
S.V.Antonyuk,
A.Marina,
S.M.Van Liempd,
S.C.Lu,
J.M.Mato,
S.S.Hasnain,
A.L.Rojas.
Structure and Function Study of the Complex That Synthesizes S-Adenosylmethionine. Iucrj V. 1 240 2014.
ISSN: ESSN 2052-2525
PubMed: 25075345
DOI: 10.1107/S2052252514012585
Page generated: Mon Aug 19 23:32:28 2024
ISSN: ESSN 2052-2525
PubMed: 25075345
DOI: 10.1107/S2052252514012585
Last articles
Cl in 7SXLCl in 7SZ8
Cl in 7SXQ
Cl in 7SWS
Cl in 7SX6
Cl in 7STD
Cl in 7SWR
Cl in 7SVT
Cl in 7SUU
Cl in 7SUJ