Magnesium »
PDB 4p9d-4pjj »
4pcj »
Magnesium in PDB 4pcj: Modifications to Toxic Cug Rnas Induce Structural Stability and Rescue Mis-Splicing in Myotonic Dystrophy
Protein crystallography data
The structure of Modifications to Toxic Cug Rnas Induce Structural Stability and Rescue Mis-Splicing in Myotonic Dystrophy, PDB code: 4pcj
was solved by
L.A.Coonrod,
E.E.Reister,
J.A.Berglund,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 22.60 / 1.90 |
| Space group | H 3 |
| Cell size a, b, c (Å), α, β, γ (°) | 69.705, 69.705, 67.797, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 19.6 / 26.8 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Modifications to Toxic Cug Rnas Induce Structural Stability and Rescue Mis-Splicing in Myotonic Dystrophy
(pdb code 4pcj). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 5 binding sites of Magnesium where determined in the Modifications to Toxic Cug Rnas Induce Structural Stability and Rescue Mis-Splicing in Myotonic Dystrophy, PDB code: 4pcj:
Jump to Magnesium binding site number: 1; 2; 3; 4; 5;
In total 5 binding sites of Magnesium where determined in the Modifications to Toxic Cug Rnas Induce Structural Stability and Rescue Mis-Splicing in Myotonic Dystrophy, PDB code: 4pcj:
Jump to Magnesium binding site number: 1; 2; 3; 4; 5;
Magnesium binding site 1 out of 5 in 4pcj
Go back to
Magnesium binding site 1 out
of 5 in the Modifications to Toxic Cug Rnas Induce Structural Stability and Rescue Mis-Splicing in Myotonic Dystrophy
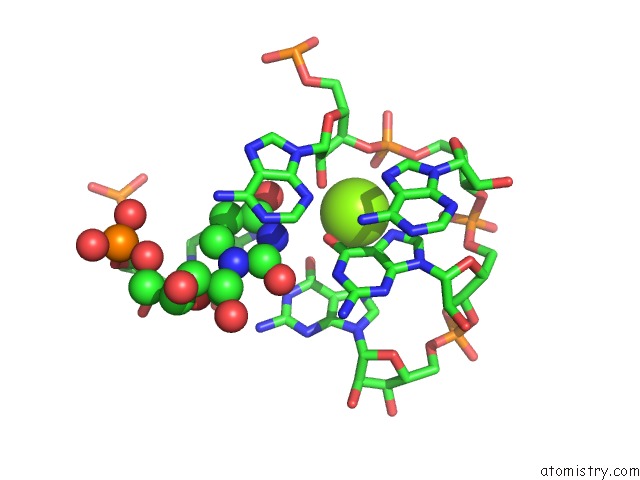
Mono view
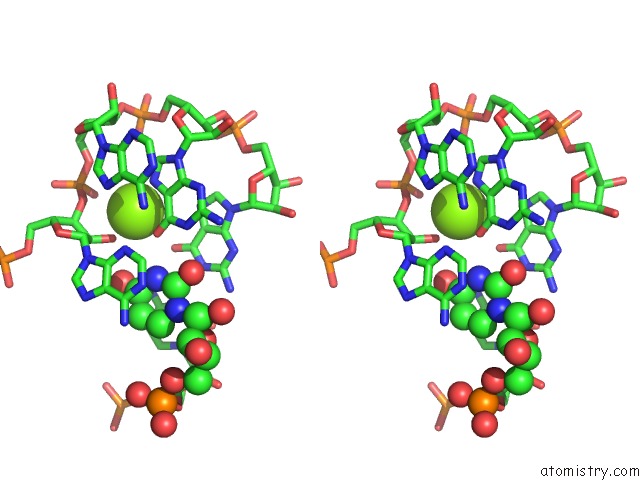
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Modifications to Toxic Cug Rnas Induce Structural Stability and Rescue Mis-Splicing in Myotonic Dystrophy within 5.0Å range:
|
Magnesium binding site 2 out of 5 in 4pcj
Go back to
Magnesium binding site 2 out
of 5 in the Modifications to Toxic Cug Rnas Induce Structural Stability and Rescue Mis-Splicing in Myotonic Dystrophy
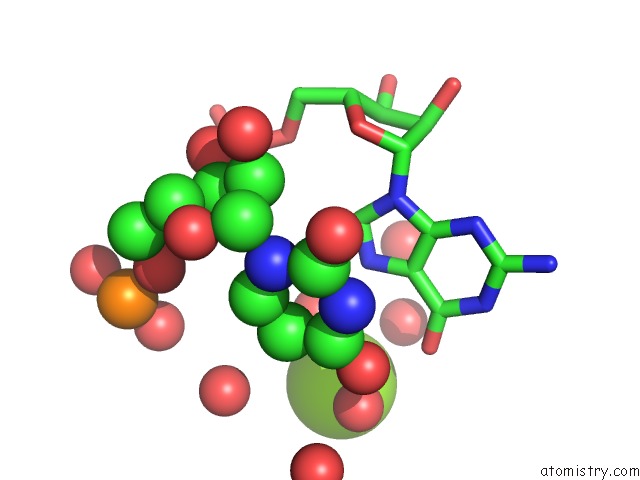
Mono view
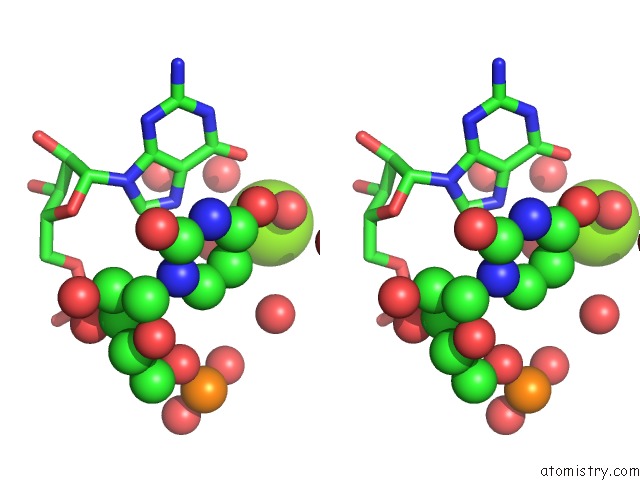
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Modifications to Toxic Cug Rnas Induce Structural Stability and Rescue Mis-Splicing in Myotonic Dystrophy within 5.0Å range:
|
Magnesium binding site 3 out of 5 in 4pcj
Go back to
Magnesium binding site 3 out
of 5 in the Modifications to Toxic Cug Rnas Induce Structural Stability and Rescue Mis-Splicing in Myotonic Dystrophy
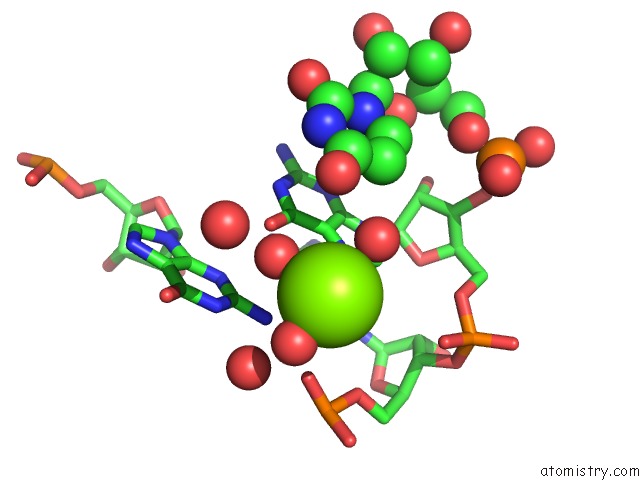
Mono view
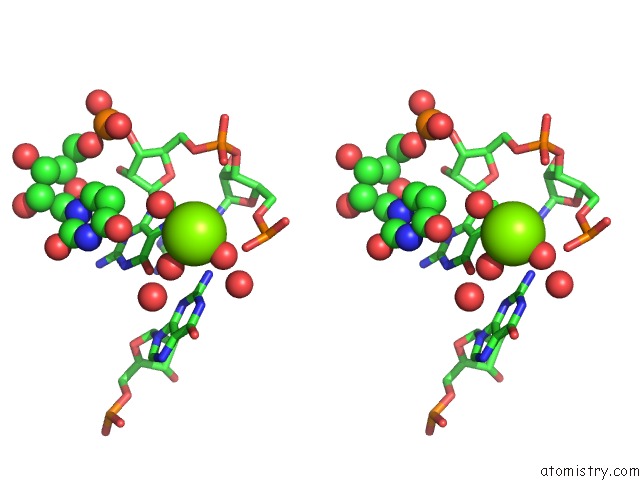
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 3 of Modifications to Toxic Cug Rnas Induce Structural Stability and Rescue Mis-Splicing in Myotonic Dystrophy within 5.0Å range:
|
Magnesium binding site 4 out of 5 in 4pcj
Go back to
Magnesium binding site 4 out
of 5 in the Modifications to Toxic Cug Rnas Induce Structural Stability and Rescue Mis-Splicing in Myotonic Dystrophy
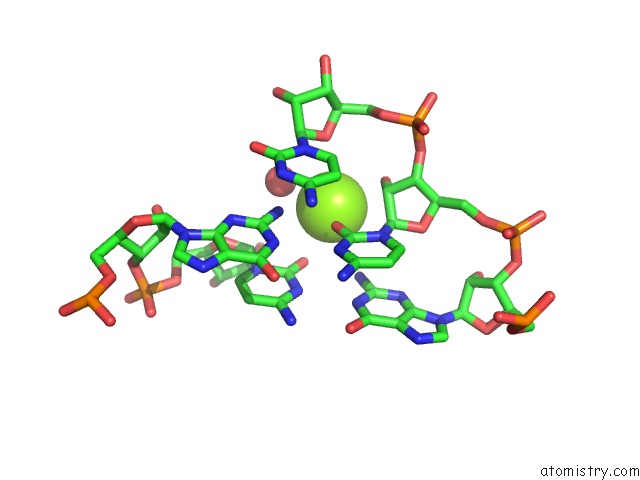
Mono view
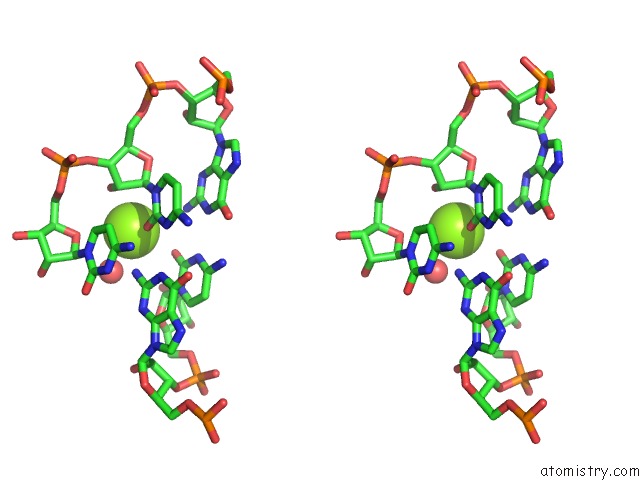
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 4 of Modifications to Toxic Cug Rnas Induce Structural Stability and Rescue Mis-Splicing in Myotonic Dystrophy within 5.0Å range:
|
Magnesium binding site 5 out of 5 in 4pcj
Go back to
Magnesium binding site 5 out
of 5 in the Modifications to Toxic Cug Rnas Induce Structural Stability and Rescue Mis-Splicing in Myotonic Dystrophy
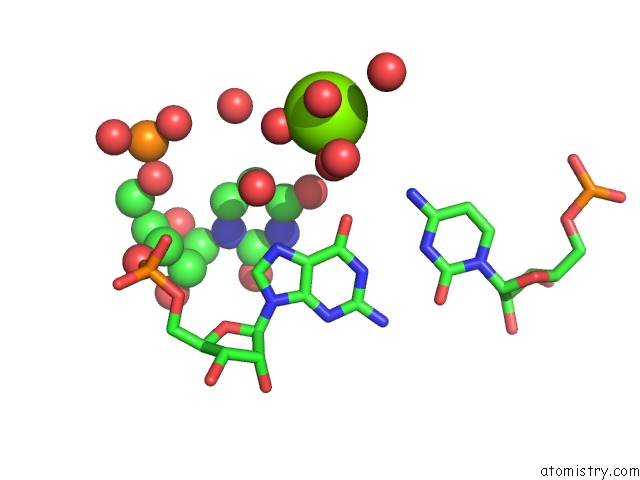
Mono view
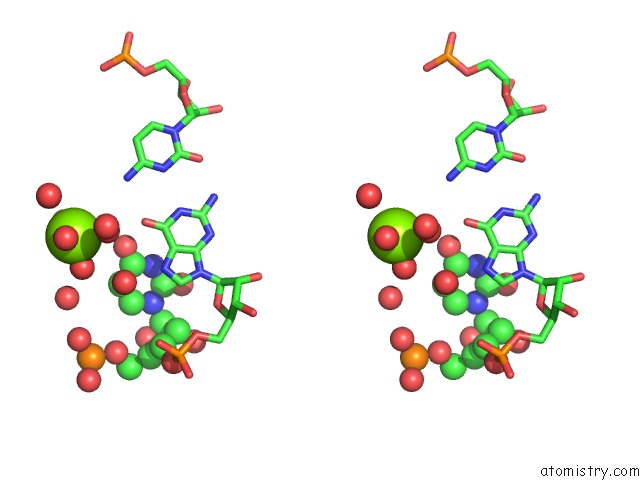
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 5 of Modifications to Toxic Cug Rnas Induce Structural Stability and Rescue Mis-Splicing in Myotonic Dystrophy within 5.0Å range:
|
Reference:
E.Delorimier,
L.A.Coonrod,
J.Copperman,
A.Taber,
E.E.Reister,
K.Sharma,
P.K.Todd,
M.G.Guenza,
J.A.Berglund.
Modifications to Toxic Cug Rnas Induce Structural Stability, Rescue Mis-Splicing in A Myotonic Dystrophy Cell Model and Reduce Toxicity in A Myotonic Dystrophy Zebrafish Model. Nucleic Acids Res. 2014.
ISSN: ESSN 1362-4962
PubMed: 25303993
DOI: 10.1093/NAR/GKU941
Page generated: Mon Aug 11 21:51:33 2025
ISSN: ESSN 1362-4962
PubMed: 25303993
DOI: 10.1093/NAR/GKU941
Last articles
Mg in 5FRNMg in 5FRM
Mg in 5FPH
Mg in 5FR2
Mg in 5FQ8
Mg in 5FR1
Mg in 5FQO
Mg in 5FQ7
Mg in 5FQ5
Mg in 5FP3