Magnesium »
PDB 4pjk-4pyk »
4puq »
Magnesium in PDB 4puq: Mus Musculus TDP2 Reaction Product Complex with 5'-Phosphorylated Rna/Dna, Glycerol, and MG2+
Protein crystallography data
The structure of Mus Musculus TDP2 Reaction Product Complex with 5'-Phosphorylated Rna/Dna, Glycerol, and MG2+, PDB code: 4puq
was solved by
M.J.Schellenberg,
R.S.Williams,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 34.03 / 1.60 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 54.828, 69.258, 167.107, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 12.9 / 17.1 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Mus Musculus TDP2 Reaction Product Complex with 5'-Phosphorylated Rna/Dna, Glycerol, and MG2+
(pdb code 4puq). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 2 binding sites of Magnesium where determined in the Mus Musculus TDP2 Reaction Product Complex with 5'-Phosphorylated Rna/Dna, Glycerol, and MG2+, PDB code: 4puq:
Jump to Magnesium binding site number: 1; 2;
In total 2 binding sites of Magnesium where determined in the Mus Musculus TDP2 Reaction Product Complex with 5'-Phosphorylated Rna/Dna, Glycerol, and MG2+, PDB code: 4puq:
Jump to Magnesium binding site number: 1; 2;
Magnesium binding site 1 out of 2 in 4puq
Go back to
Magnesium binding site 1 out
of 2 in the Mus Musculus TDP2 Reaction Product Complex with 5'-Phosphorylated Rna/Dna, Glycerol, and MG2+
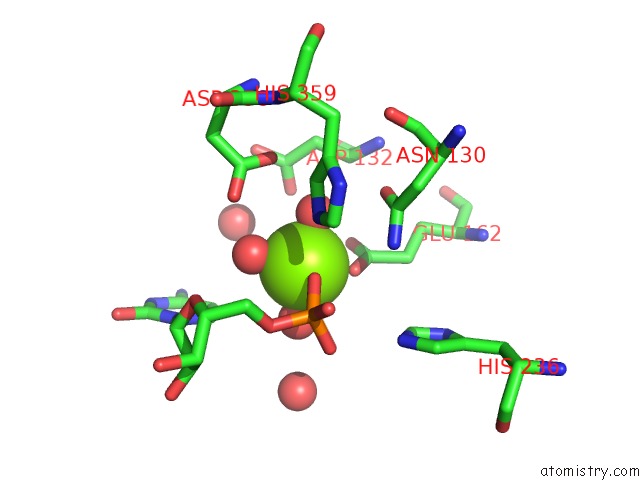
Mono view
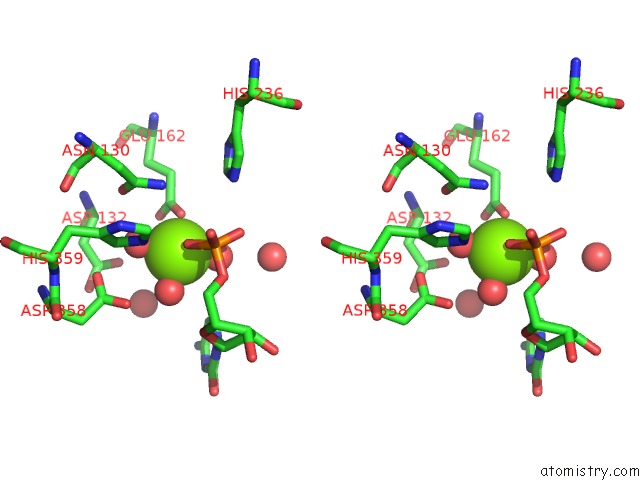
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Mus Musculus TDP2 Reaction Product Complex with 5'-Phosphorylated Rna/Dna, Glycerol, and MG2+ within 5.0Å range:
|
Magnesium binding site 2 out of 2 in 4puq
Go back to
Magnesium binding site 2 out
of 2 in the Mus Musculus TDP2 Reaction Product Complex with 5'-Phosphorylated Rna/Dna, Glycerol, and MG2+
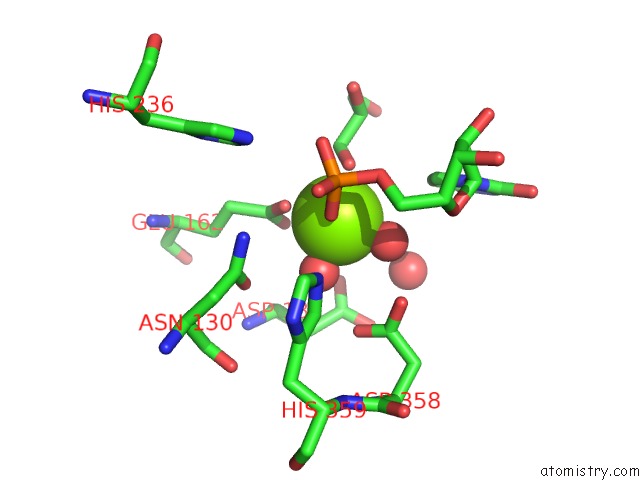
Mono view
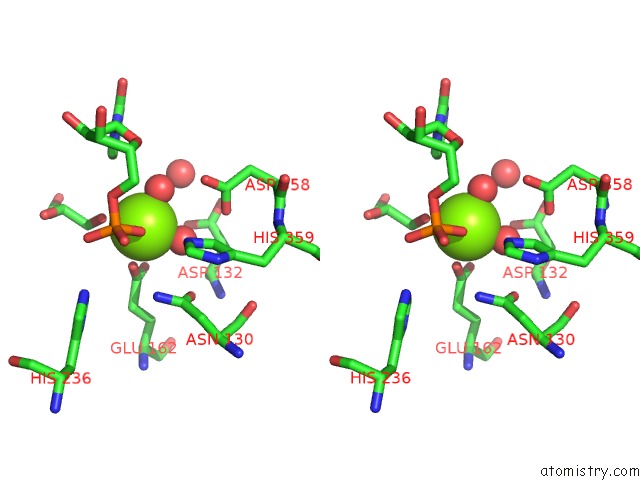
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Mus Musculus TDP2 Reaction Product Complex with 5'-Phosphorylated Rna/Dna, Glycerol, and MG2+ within 5.0Å range:
|
Reference:
R.Gao,
M.J.Schellenberg,
S.Y.Huang,
M.Abdelmalak,
C.Marchand,
K.C.Nitiss,
J.L.Nitiss,
R.S.Williams,
Y.Pommier.
Proteolytic Degradation of Topoisomerase II (TOP2) Enables the Processing of TOP2DNA and TOP2RNA Covalent Complexes By Tyrosyl-Dna-Phosphodiesterase 2 (TDP2). J.Biol.Chem. V. 289 17960 2014.
ISSN: ISSN 0021-9258
PubMed: 24808172
DOI: 10.1074/JBC.M114.565374
Page generated: Mon Aug 11 22:10:08 2025
ISSN: ISSN 0021-9258
PubMed: 24808172
DOI: 10.1074/JBC.M114.565374
Last articles
Mg in 4X5CMg in 4X5E
Mg in 4X5V
Mg in 4X5B
Mg in 4X59
Mg in 4X58
Mg in 4X4V
Mg in 4X4S
Mg in 4X4R
Mg in 4X4Q