Magnesium »
PDB 4qhe-4qpz »
4qka »
Magnesium in PDB 4qka: C-Di-Amp Riboswitch From Thermoanaerobacter Pseudethanolicus, Iridium Hexamine Soak
Protein crystallography data
The structure of C-Di-Amp Riboswitch From Thermoanaerobacter Pseudethanolicus, Iridium Hexamine Soak, PDB code: 4qka
was solved by
A.Gao,
A.Serganov,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 29.01 / 3.20 |
| Space group | P 31 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 116.035, 116.035, 114.098, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 17.1 / 19.7 |
Other elements in 4qka:
The structure of C-Di-Amp Riboswitch From Thermoanaerobacter Pseudethanolicus, Iridium Hexamine Soak also contains other interesting chemical elements:
| Potassium | (K) | 1 atom |
| Iridium | (Ir) | 5 atoms |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the C-Di-Amp Riboswitch From Thermoanaerobacter Pseudethanolicus, Iridium Hexamine Soak
(pdb code 4qka). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total only one binding site of Magnesium was determined in the C-Di-Amp Riboswitch From Thermoanaerobacter Pseudethanolicus, Iridium Hexamine Soak, PDB code: 4qka:
In total only one binding site of Magnesium was determined in the C-Di-Amp Riboswitch From Thermoanaerobacter Pseudethanolicus, Iridium Hexamine Soak, PDB code: 4qka:
Magnesium binding site 1 out of 1 in 4qka
Go back to
Magnesium binding site 1 out
of 1 in the C-Di-Amp Riboswitch From Thermoanaerobacter Pseudethanolicus, Iridium Hexamine Soak
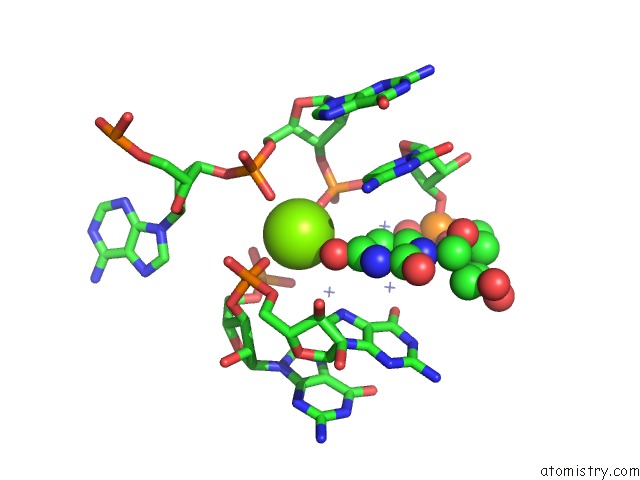
Mono view
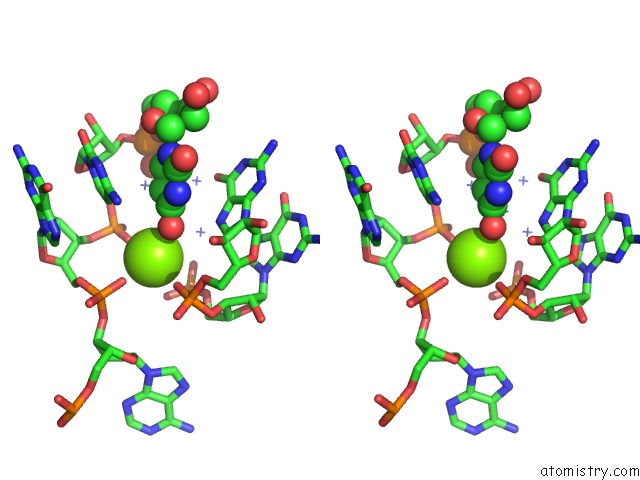
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of C-Di-Amp Riboswitch From Thermoanaerobacter Pseudethanolicus, Iridium Hexamine Soak within 5.0Å range:
|
Reference:
A.Gao,
A.Serganov.
Structural Insights Into Recognition of C-Di-Amp By the Ydao Riboswitch. Nat.Chem.Biol. V. 10 787 2014.
ISSN: ISSN 1552-4450
PubMed: 25086507
DOI: 10.1038/NCHEMBIO.1607
Page generated: Tue Aug 20 02:02:03 2024
ISSN: ISSN 1552-4450
PubMed: 25086507
DOI: 10.1038/NCHEMBIO.1607
Last articles
Ca in 5VLKCa in 5VLH
Ca in 5VLA
Ca in 5VKW
Ca in 5VF4
Ca in 5VJF
Ca in 5VGC
Ca in 5VGT
Ca in 5VER
Ca in 5VEQ