Magnesium »
PDB 4rkq-4rri »
4rnk »
Magnesium in PDB 4rnk: Sequence and Structure of A Self-Assembled 3-D Dna Crystal: D(Ggaaaatttggag)
Protein crystallography data
The structure of Sequence and Structure of A Self-Assembled 3-D Dna Crystal: D(Ggaaaatttggag), PDB code: 4rnk
was solved by
M.M.Saoji,
P.J.Paukstelis,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 34.41 / 2.08 |
| Space group | P 64 |
| Cell size a, b, c (Å), α, β, γ (°) | 39.729, 39.729, 55.989, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 20.1 / 23.9 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Sequence and Structure of A Self-Assembled 3-D Dna Crystal: D(Ggaaaatttggag)
(pdb code 4rnk). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 2 binding sites of Magnesium where determined in the Sequence and Structure of A Self-Assembled 3-D Dna Crystal: D(Ggaaaatttggag), PDB code: 4rnk:
Jump to Magnesium binding site number: 1; 2;
In total 2 binding sites of Magnesium where determined in the Sequence and Structure of A Self-Assembled 3-D Dna Crystal: D(Ggaaaatttggag), PDB code: 4rnk:
Jump to Magnesium binding site number: 1; 2;
Magnesium binding site 1 out of 2 in 4rnk
Go back to
Magnesium binding site 1 out
of 2 in the Sequence and Structure of A Self-Assembled 3-D Dna Crystal: D(Ggaaaatttggag)
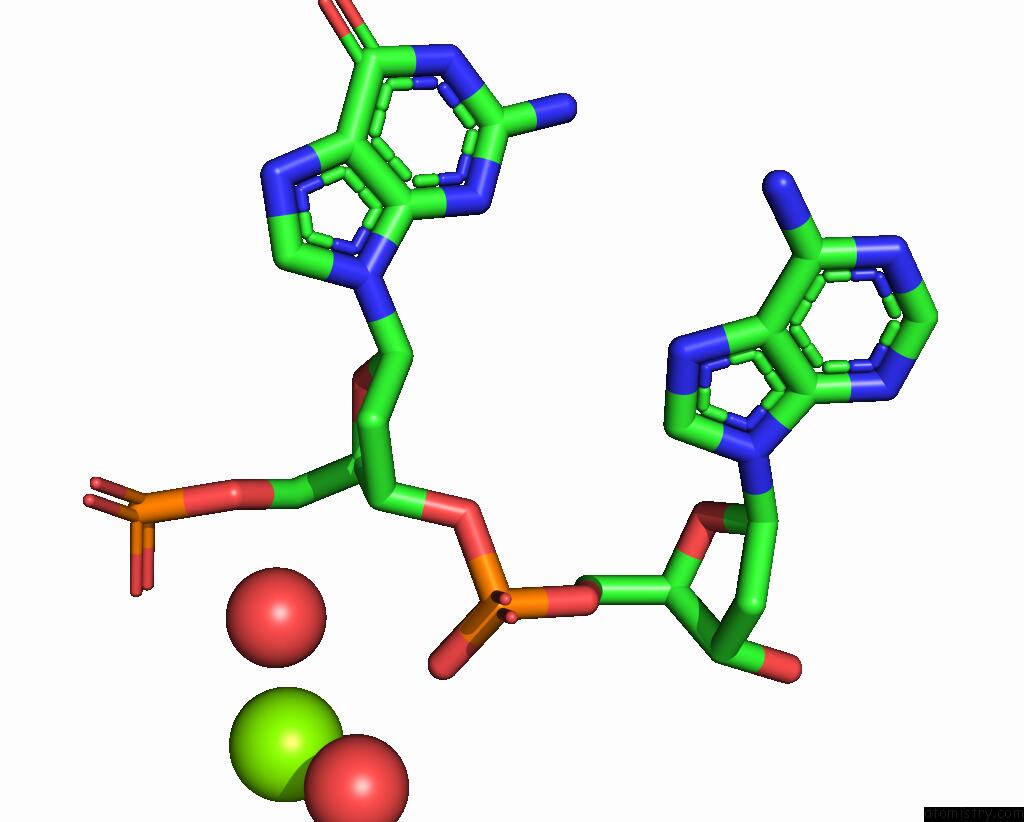
Mono view
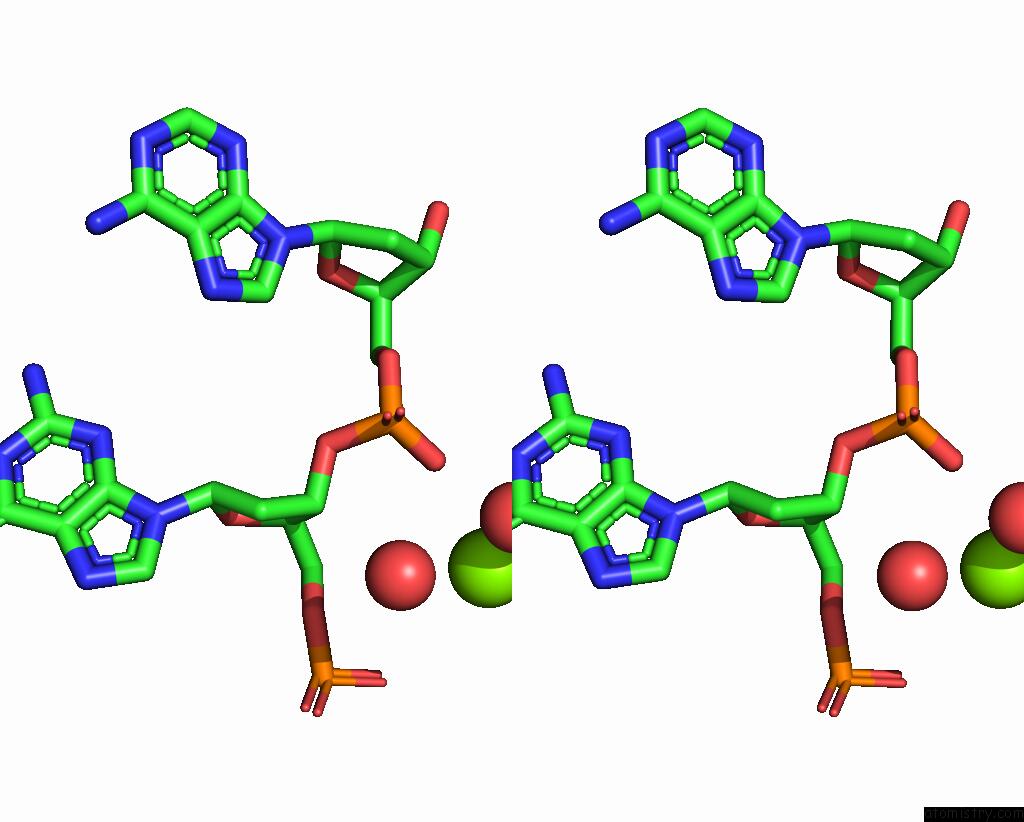
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Sequence and Structure of A Self-Assembled 3-D Dna Crystal: D(Ggaaaatttggag) within 5.0Å range:
|
Magnesium binding site 2 out of 2 in 4rnk
Go back to
Magnesium binding site 2 out
of 2 in the Sequence and Structure of A Self-Assembled 3-D Dna Crystal: D(Ggaaaatttggag)
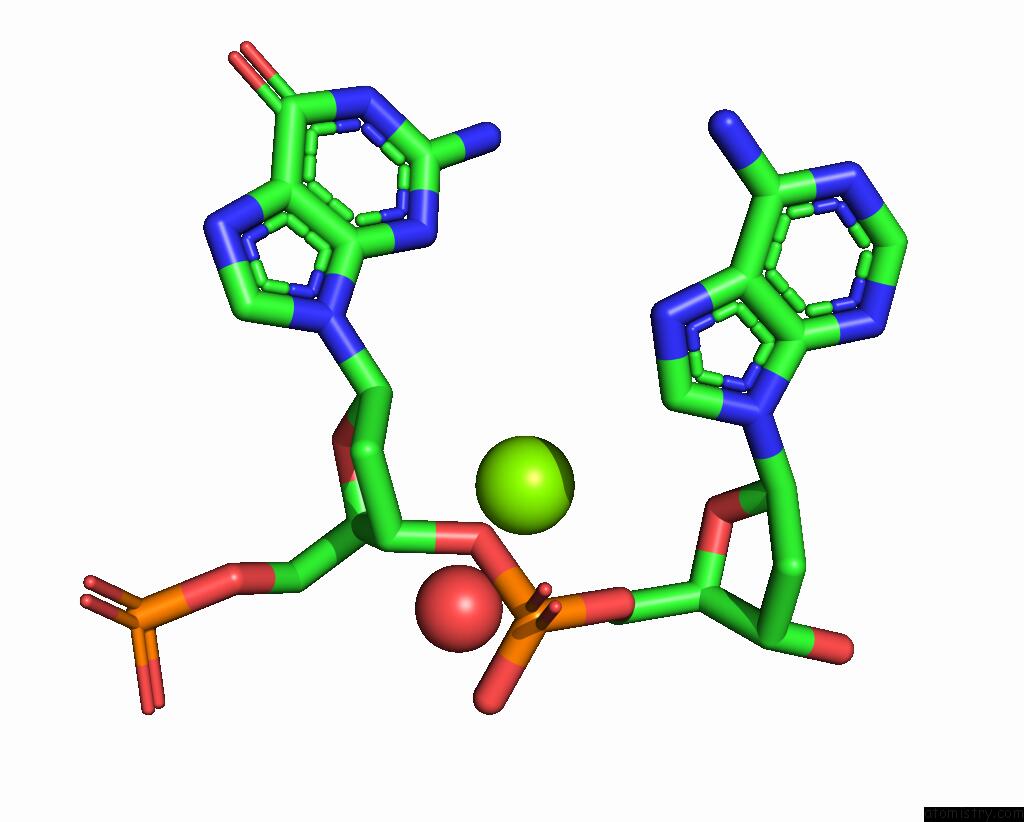
Mono view
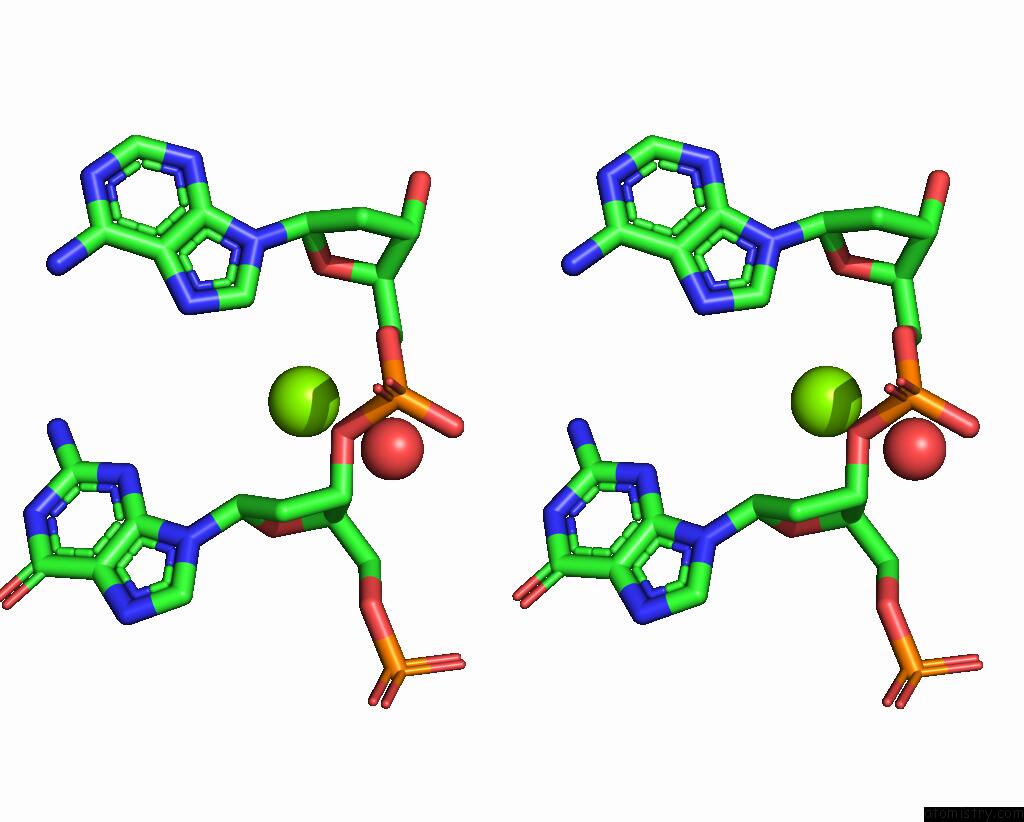
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Sequence and Structure of A Self-Assembled 3-D Dna Crystal: D(Ggaaaatttggag) within 5.0Å range:
|
Reference:
M.Saoji,
D.Zhang,
P.J.Paukstelis.
Probing the Role of Sequence in the Assembly of Three-Dimensional Dna Crystals. Biopolymers V. 103 618 2015.
ISSN: ISSN 0006-3525
PubMed: 26015367
DOI: 10.1002/BIP.22688
Page generated: Tue Aug 20 03:13:41 2024
ISSN: ISSN 0006-3525
PubMed: 26015367
DOI: 10.1002/BIP.22688
Last articles
Al in 8SGCAl in 8SG9
Al in 8SFF
Al in 8SG8
Al in 8R1A
Al in 8Q75
Al in 8OIE
Al in 8OX6
Al in 8OX5
Al in 8OP8