Magnesium »
PDB 4s1m-4tnz »
4tnp »
Magnesium in PDB 4tnp: Structural Basis of Cellular Dntp Regulation, SAMHD1-Gtp-Dctp-Cctp Complex
Protein crystallography data
The structure of Structural Basis of Cellular Dntp Regulation, SAMHD1-Gtp-Dctp-Cctp Complex, PDB code: 4tnp
was solved by
X.Ji,
C.Tang,
Q.Zhao,
W.Wang,
Y.Xiong,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 50.00 / 2.00 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 81.507, 140.753, 97.490, 90.00, 114.79, 90.00 |
| R / Rfree (%) | 19.8 / 22.2 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Structural Basis of Cellular Dntp Regulation, SAMHD1-Gtp-Dctp-Cctp Complex
(pdb code 4tnp). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 4 binding sites of Magnesium where determined in the Structural Basis of Cellular Dntp Regulation, SAMHD1-Gtp-Dctp-Cctp Complex, PDB code: 4tnp:
Jump to Magnesium binding site number: 1; 2; 3; 4;
In total 4 binding sites of Magnesium where determined in the Structural Basis of Cellular Dntp Regulation, SAMHD1-Gtp-Dctp-Cctp Complex, PDB code: 4tnp:
Jump to Magnesium binding site number: 1; 2; 3; 4;
Magnesium binding site 1 out of 4 in 4tnp
Go back to
Magnesium binding site 1 out
of 4 in the Structural Basis of Cellular Dntp Regulation, SAMHD1-Gtp-Dctp-Cctp Complex
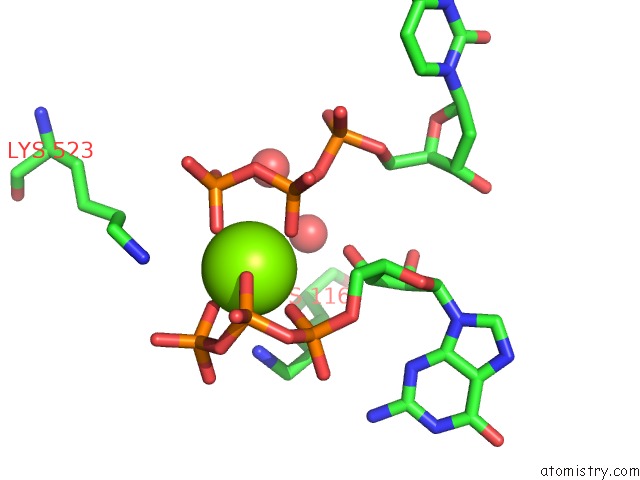
Mono view
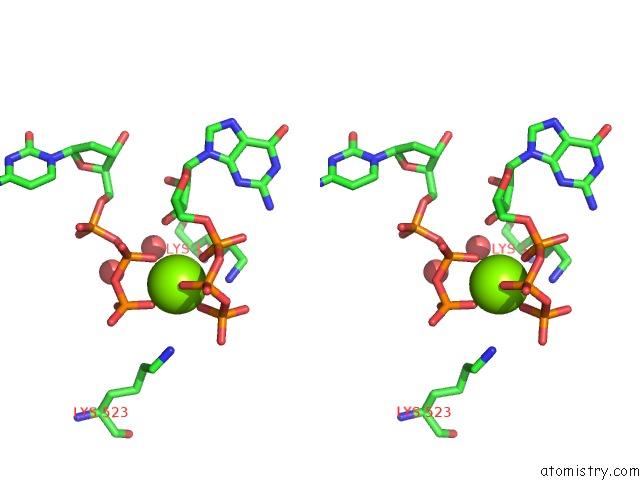
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Structural Basis of Cellular Dntp Regulation, SAMHD1-Gtp-Dctp-Cctp Complex within 5.0Å range:
|
Magnesium binding site 2 out of 4 in 4tnp
Go back to
Magnesium binding site 2 out
of 4 in the Structural Basis of Cellular Dntp Regulation, SAMHD1-Gtp-Dctp-Cctp Complex
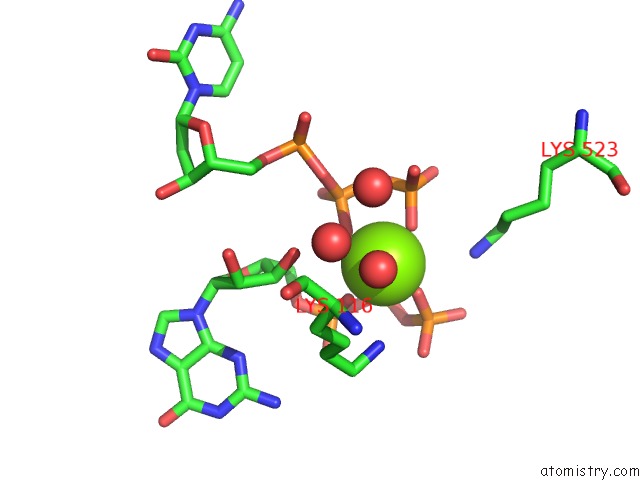
Mono view
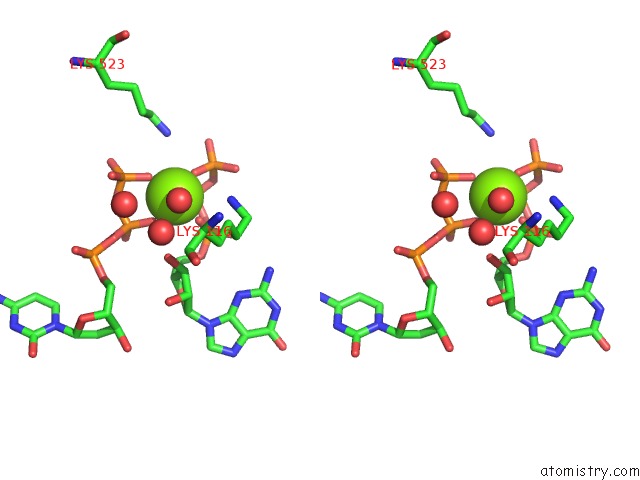
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Structural Basis of Cellular Dntp Regulation, SAMHD1-Gtp-Dctp-Cctp Complex within 5.0Å range:
|
Magnesium binding site 3 out of 4 in 4tnp
Go back to
Magnesium binding site 3 out
of 4 in the Structural Basis of Cellular Dntp Regulation, SAMHD1-Gtp-Dctp-Cctp Complex
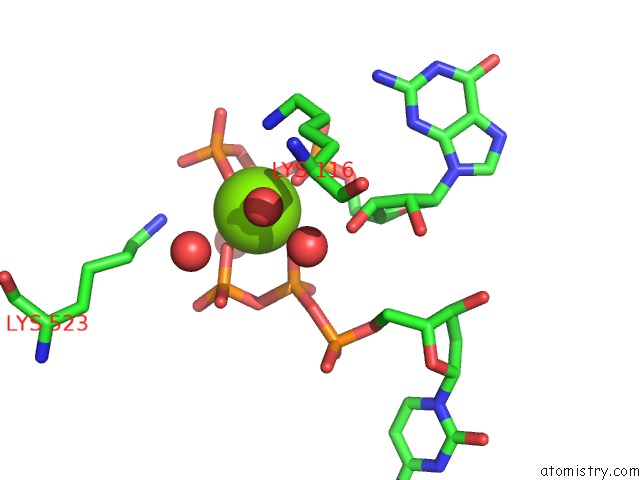
Mono view
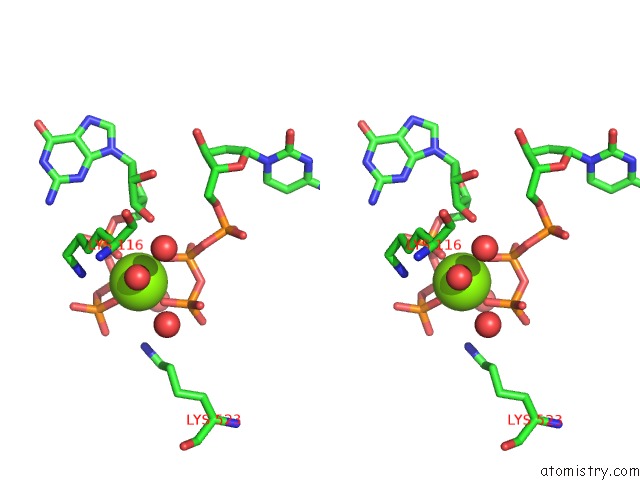
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 3 of Structural Basis of Cellular Dntp Regulation, SAMHD1-Gtp-Dctp-Cctp Complex within 5.0Å range:
|
Magnesium binding site 4 out of 4 in 4tnp
Go back to
Magnesium binding site 4 out
of 4 in the Structural Basis of Cellular Dntp Regulation, SAMHD1-Gtp-Dctp-Cctp Complex
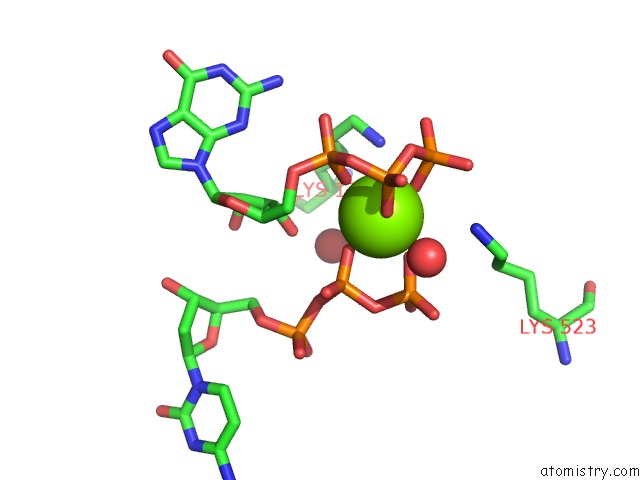
Mono view
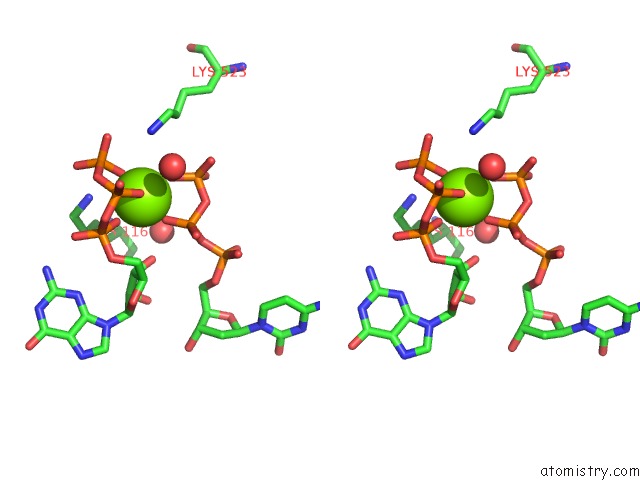
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 4 of Structural Basis of Cellular Dntp Regulation, SAMHD1-Gtp-Dctp-Cctp Complex within 5.0Å range:
|
Reference:
X.Ji,
C.Tang,
Q.Zhao,
W.Wang,
Y.Xiong.
Structural Basis of Cellular Dntp Regulation By SAMHD1. Proc.Natl.Acad.Sci.Usa V. 111 E4305 2014.
ISSN: ESSN 1091-6490
PubMed: 25267621
DOI: 10.1073/PNAS.1412289111
Page generated: Tue Aug 20 04:07:08 2024
ISSN: ESSN 1091-6490
PubMed: 25267621
DOI: 10.1073/PNAS.1412289111
Last articles
F in 7LCRF in 7LCM
F in 7LCO
F in 7LCK
F in 7LCJ
F in 7LCI
F in 7L9Y
F in 7LCD
F in 7LAY
F in 7LAJ