Magnesium »
PDB 4uvd-4w5o »
4uys »
Magnesium in PDB 4uys: X-Ray Structure of the N-Terminal Domain of the Flocculin FLO11 From Saccharomyces Cerevisiae
Protein crystallography data
The structure of X-Ray Structure of the N-Terminal Domain of the Flocculin FLO11 From Saccharomyces Cerevisiae, PDB code: 4uys
was solved by
T.Kraushaar,
M.Veelders,
S.Brueckner,
D.Rhinow,
H.U.Moesch,
L.O.Essen,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 28.51 / 1.05 |
| Space group | P 21 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 52.720, 101.550, 33.870, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 14.683 / 16.968 |
Other elements in 4uys:
The structure of X-Ray Structure of the N-Terminal Domain of the Flocculin FLO11 From Saccharomyces Cerevisiae also contains other interesting chemical elements:
| Sodium | (Na) | 7 atoms |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the X-Ray Structure of the N-Terminal Domain of the Flocculin FLO11 From Saccharomyces Cerevisiae
(pdb code 4uys). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total only one binding site of Magnesium was determined in the X-Ray Structure of the N-Terminal Domain of the Flocculin FLO11 From Saccharomyces Cerevisiae, PDB code: 4uys:
In total only one binding site of Magnesium was determined in the X-Ray Structure of the N-Terminal Domain of the Flocculin FLO11 From Saccharomyces Cerevisiae, PDB code: 4uys:
Magnesium binding site 1 out of 1 in 4uys
Go back to
Magnesium binding site 1 out
of 1 in the X-Ray Structure of the N-Terminal Domain of the Flocculin FLO11 From Saccharomyces Cerevisiae
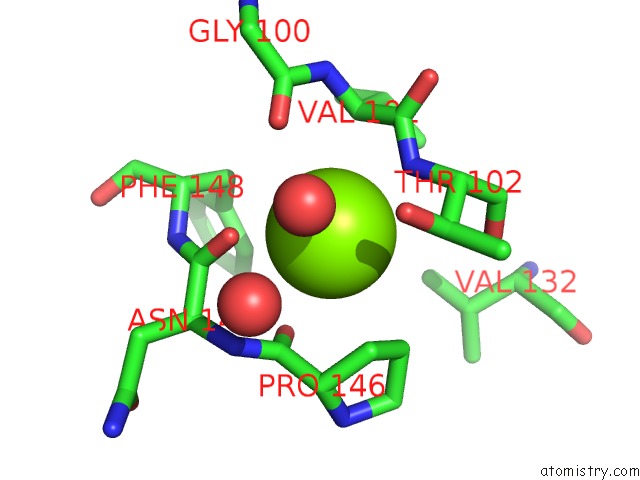
Mono view
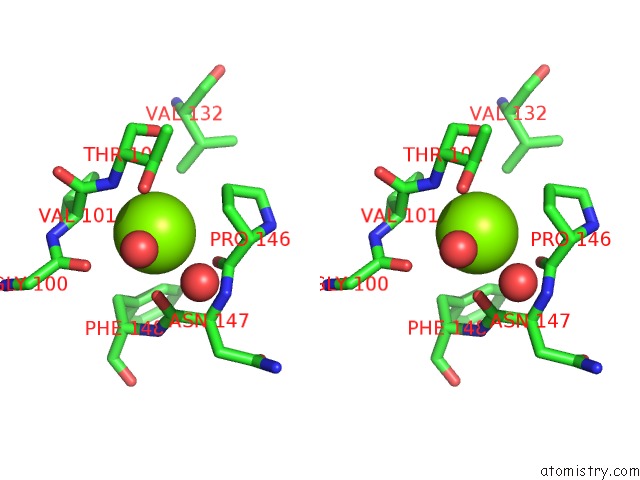
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of X-Ray Structure of the N-Terminal Domain of the Flocculin FLO11 From Saccharomyces Cerevisiae within 5.0Å range:
|
Reference:
T.Kraushaar,
S.Bruckner,
M.Veelders,
D.Rhinow,
F.Schreiner,
R.Birke,
A.Pagenstecher,
H.Mosch,
L.Essen.
Interactions By the Fungal FLO11 Adhesin Depend on A Fibronectin Type III-Like Adhesin Domain Girdled By Aromatic Bands. Structure V. 23 1005 2015.
ISSN: ISSN 0969-2126
PubMed: 25960408
DOI: 10.1016/J.STR.2015.03.021
Page generated: Tue Aug 12 00:49:31 2025
ISSN: ISSN 0969-2126
PubMed: 25960408
DOI: 10.1016/J.STR.2015.03.021
Last articles
Mg in 5C1TMg in 5C1O
Mg in 5C12
Mg in 5C0P
Mg in 5C0W
Mg in 5C0Y
Mg in 5C03
Mg in 5BZY
Mg in 5BYL
Mg in 5BZ9