Magnesium »
PDB 4xtr-4y2q »
4xz3 »
Magnesium in PDB 4xz3: Ca. Korarchaeum Cryptofilum Dinucleotide Forming Acetyl-Coenzyme A Synthetase 1 (Se-Met Derivative) in Complex with Coenzyme A and Mg- Amppcp, Phosphohistidine Segment Pointing Towards Nucleotide Binding Site
Protein crystallography data
The structure of Ca. Korarchaeum Cryptofilum Dinucleotide Forming Acetyl-Coenzyme A Synthetase 1 (Se-Met Derivative) in Complex with Coenzyme A and Mg- Amppcp, Phosphohistidine Segment Pointing Towards Nucleotide Binding Site, PDB code: 4xz3
was solved by
R.H.-J.Weisse,
A.J.Scheidig,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 48.84 / 2.40 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 100.170, 111.880, 127.590, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 19.7 / 24.4 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Ca. Korarchaeum Cryptofilum Dinucleotide Forming Acetyl-Coenzyme A Synthetase 1 (Se-Met Derivative) in Complex with Coenzyme A and Mg- Amppcp, Phosphohistidine Segment Pointing Towards Nucleotide Binding Site
(pdb code 4xz3). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 5 binding sites of Magnesium where determined in the Ca. Korarchaeum Cryptofilum Dinucleotide Forming Acetyl-Coenzyme A Synthetase 1 (Se-Met Derivative) in Complex with Coenzyme A and Mg- Amppcp, Phosphohistidine Segment Pointing Towards Nucleotide Binding Site, PDB code: 4xz3:
Jump to Magnesium binding site number: 1; 2; 3; 4; 5;
In total 5 binding sites of Magnesium where determined in the Ca. Korarchaeum Cryptofilum Dinucleotide Forming Acetyl-Coenzyme A Synthetase 1 (Se-Met Derivative) in Complex with Coenzyme A and Mg- Amppcp, Phosphohistidine Segment Pointing Towards Nucleotide Binding Site, PDB code: 4xz3:
Jump to Magnesium binding site number: 1; 2; 3; 4; 5;
Magnesium binding site 1 out of 5 in 4xz3
Go back to
Magnesium binding site 1 out
of 5 in the Ca. Korarchaeum Cryptofilum Dinucleotide Forming Acetyl-Coenzyme A Synthetase 1 (Se-Met Derivative) in Complex with Coenzyme A and Mg- Amppcp, Phosphohistidine Segment Pointing Towards Nucleotide Binding Site
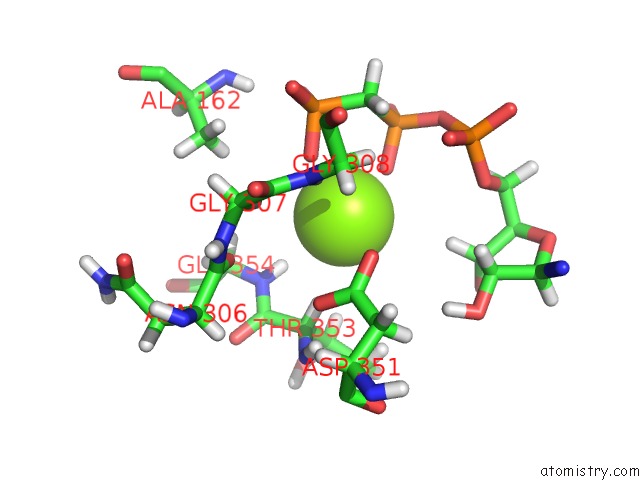
Mono view
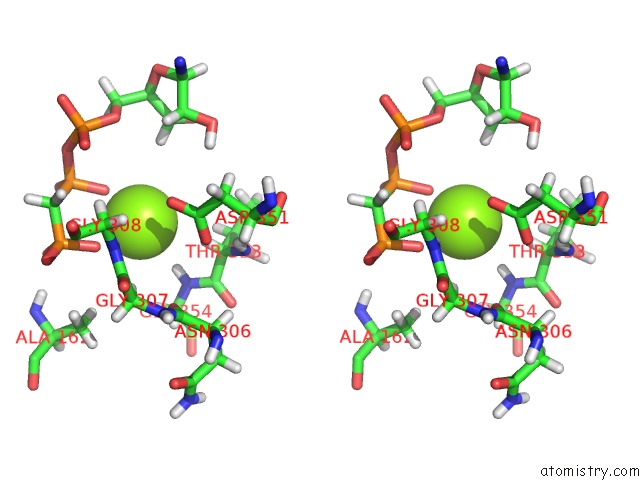
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Ca. Korarchaeum Cryptofilum Dinucleotide Forming Acetyl-Coenzyme A Synthetase 1 (Se-Met Derivative) in Complex with Coenzyme A and Mg- Amppcp, Phosphohistidine Segment Pointing Towards Nucleotide Binding Site within 5.0Å range:
|
Magnesium binding site 2 out of 5 in 4xz3
Go back to
Magnesium binding site 2 out
of 5 in the Ca. Korarchaeum Cryptofilum Dinucleotide Forming Acetyl-Coenzyme A Synthetase 1 (Se-Met Derivative) in Complex with Coenzyme A and Mg- Amppcp, Phosphohistidine Segment Pointing Towards Nucleotide Binding Site
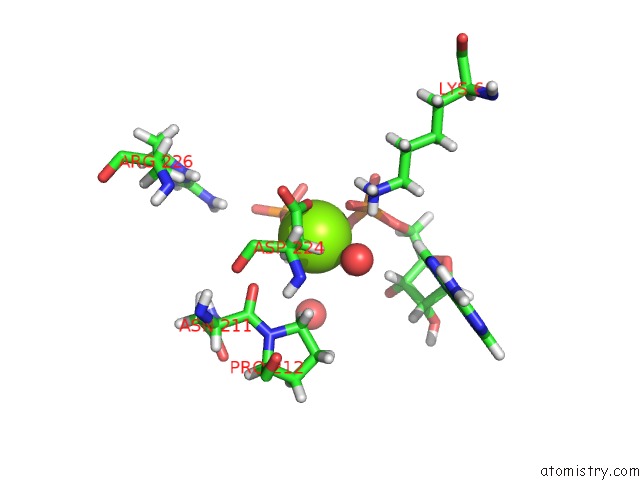
Mono view
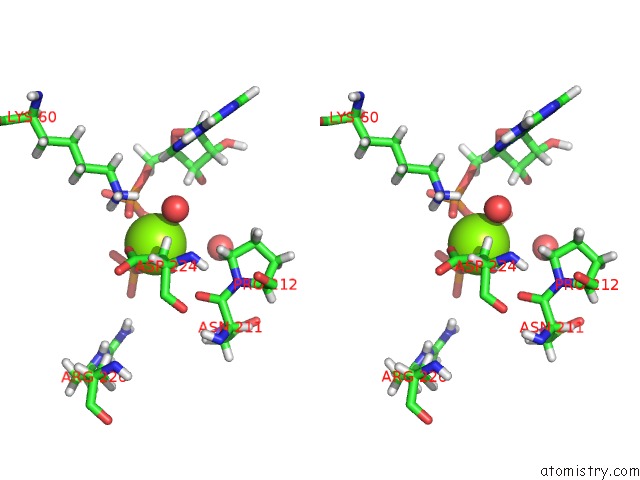
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Ca. Korarchaeum Cryptofilum Dinucleotide Forming Acetyl-Coenzyme A Synthetase 1 (Se-Met Derivative) in Complex with Coenzyme A and Mg- Amppcp, Phosphohistidine Segment Pointing Towards Nucleotide Binding Site within 5.0Å range:
|
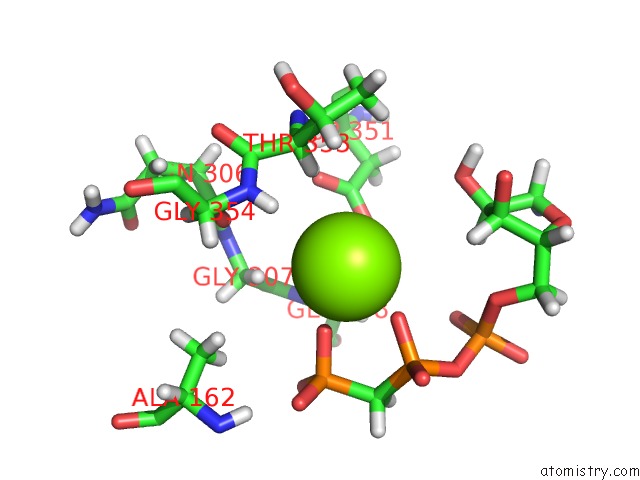
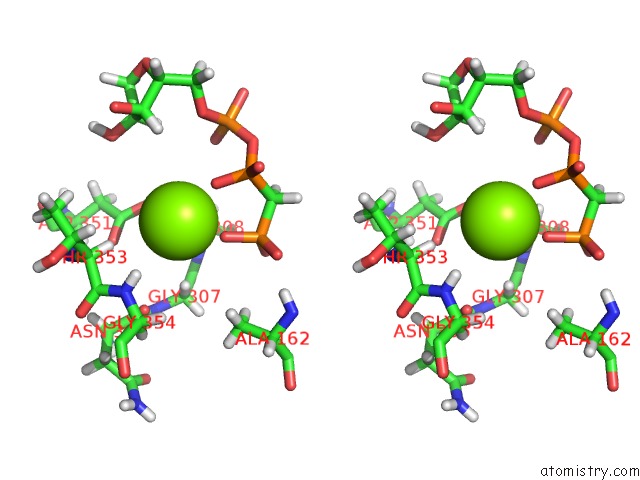
Magnesium binding site 3 out of 5 in 4xz3
Go back to
Magnesium binding site 3 out
of 5 in the Ca. Korarchaeum Cryptofilum Dinucleotide Forming Acetyl-Coenzyme A Synthetase 1 (Se-Met Derivative) in Complex with Coenzyme A and Mg- Amppcp, Phosphohistidine Segment Pointing Towards Nucleotide Binding Site
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 3 of Ca. Korarchaeum Cryptofilum Dinucleotide Forming Acetyl-Coenzyme A Synthetase 1 (Se-Met Derivative) in Complex with Coenzyme A and Mg- Amppcp, Phosphohistidine Segment Pointing Towards Nucleotide Binding Site within 5.0Å range:
|
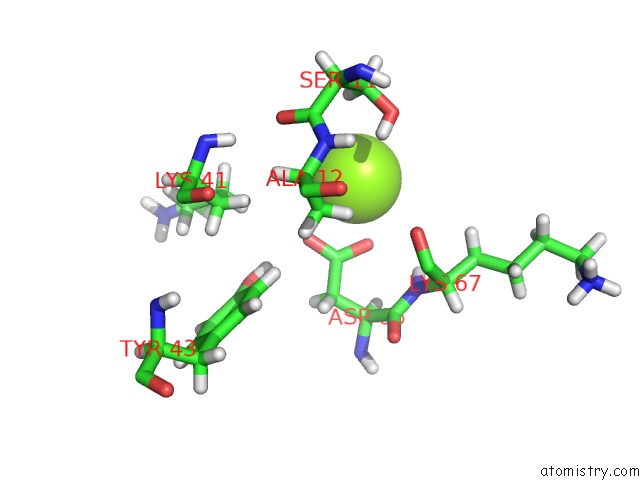
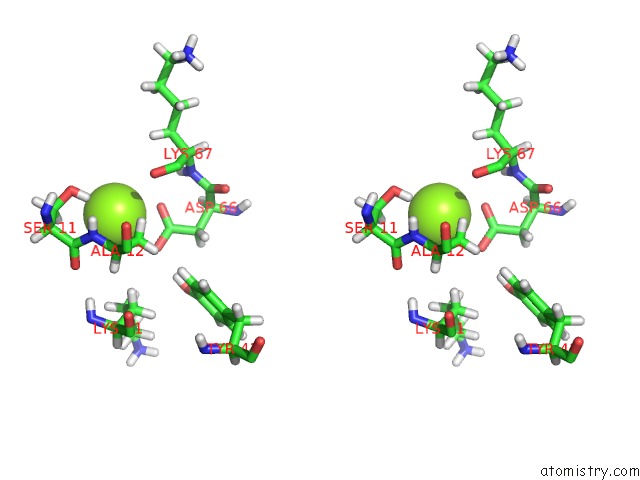
Magnesium binding site 4 out of 5 in 4xz3
Go back to
Magnesium binding site 4 out
of 5 in the Ca. Korarchaeum Cryptofilum Dinucleotide Forming Acetyl-Coenzyme A Synthetase 1 (Se-Met Derivative) in Complex with Coenzyme A and Mg- Amppcp, Phosphohistidine Segment Pointing Towards Nucleotide Binding Site
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 4 of Ca. Korarchaeum Cryptofilum Dinucleotide Forming Acetyl-Coenzyme A Synthetase 1 (Se-Met Derivative) in Complex with Coenzyme A and Mg- Amppcp, Phosphohistidine Segment Pointing Towards Nucleotide Binding Site within 5.0Å range:
|
Magnesium binding site 5 out of 5 in 4xz3
Go back to
Magnesium binding site 5 out
of 5 in the Ca. Korarchaeum Cryptofilum Dinucleotide Forming Acetyl-Coenzyme A Synthetase 1 (Se-Met Derivative) in Complex with Coenzyme A and Mg- Amppcp, Phosphohistidine Segment Pointing Towards Nucleotide Binding Site
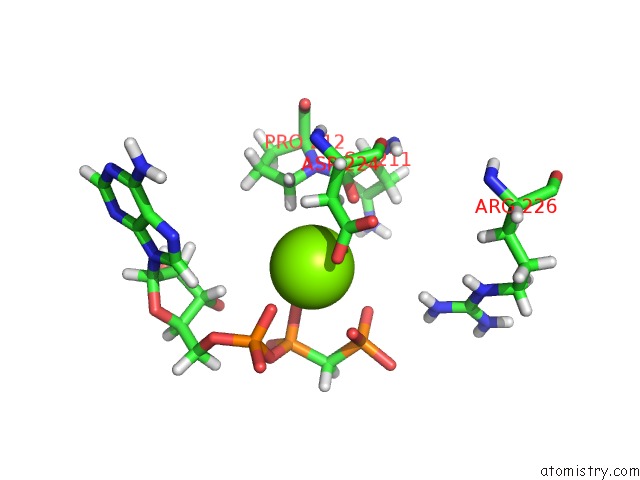
Mono view
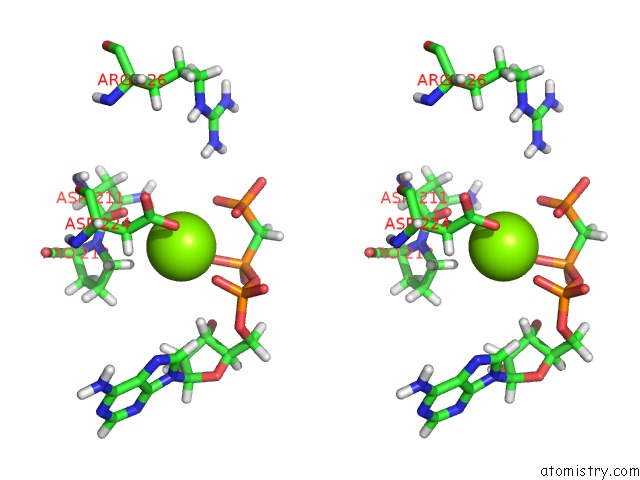
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 5 of Ca. Korarchaeum Cryptofilum Dinucleotide Forming Acetyl-Coenzyme A Synthetase 1 (Se-Met Derivative) in Complex with Coenzyme A and Mg- Amppcp, Phosphohistidine Segment Pointing Towards Nucleotide Binding Site within 5.0Å range:
|
Reference:
R.H.Weie,
A.Faust,
M.Schmidt,
P.Schonheit,
A.J.Scheidig.
Structure of Ndp-Forming Acetyl-Coa Synthetase ACD1 Reveals A Large Rearrangement For Phosphoryl Transfer. Proc.Natl.Acad.Sci.Usa V. 113 E519 2016.
ISSN: ESSN 1091-6490
PubMed: 26787904
DOI: 10.1073/PNAS.1518614113
Page generated: Tue Aug 20 16:03:54 2024
ISSN: ESSN 1091-6490
PubMed: 26787904
DOI: 10.1073/PNAS.1518614113
Last articles
Cl in 5VZHCl in 5VZB
Cl in 5W0J
Cl in 5VZG
Cl in 5VZE
Cl in 5VZC
Cl in 5VZ9
Cl in 5VZ7
Cl in 5VYY
Cl in 5VYQ